Immunotherapy response induces divergent tertiary lymphoid structure morphologies in hepatocellular carcinoma
- PMID: 39455893
- PMCID: PMC12042221
- DOI: 10.1038/s41590-024-01992-w
Immunotherapy response induces divergent tertiary lymphoid structure morphologies in hepatocellular carcinoma
Abstract
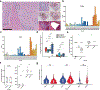
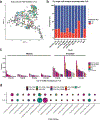
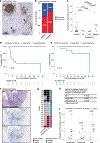
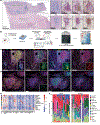
Tertiary lymphoid structures (TLS) are associated with improved response in solid tumors treated with immune checkpoint blockade, but understanding of the prognostic and predictive value of TLS and the circumstances of their resolution is incomplete. Here we show that in hepatocellular carcinoma treated with neoadjuvant immunotherapy, high intratumoral TLS density at the time of surgery is associated with pathologic response and improved relapse-free survival. In areas of tumor regression, we identify a noncanonical involuted morphology of TLS marked by dispersion of the B cell follicle, persistence of a T cell zone enriched for T cell-mature dendritic cell interactions and increased expression of T cell memory markers. Collectively, these data suggest that TLS can serve as both a prognostic and predictive marker of response to immunotherapy in hepatocellular carcinoma and that late-stage TLS may support T cell memory formation after elimination of a viable tumor.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
M.Y. reports consulting fees from Astra Zeneca, Exelixis, Genentech, Replimune, Hepion, Lantheus; research funding (to Johns Hopkins) from Bristol-Myers Squibb, Exelixis, Incyte and Genentech; and is a co-founder with equity of Adventris Pharmaceuticals. E.J.F. is on the Scientific Advisory Board of Viosera/Resistance Bio, is a paid consultant for Merck and Mestag Therapeutics, and receives research funds from Abbvie. W.J.H. has received patent royalties from Rodeo/Amgen and is the recipient of grants from Sanofi, NeoTX, and CirclePharma. He has received speaking/travel honoraria from Exelixis and Standard BioTools. E.M.J. reports grant/research support from the Lustgarten Foundation, Break Through Cancer, Roche/Genentech, Bristol-Meyers Squibb; honoraria from Achilles, DragonFly, Parker Institute, Cancer Prevention and Research Institute of Texas, Surge, HDT Bio, Mestag Therapeutics, Medical Home Group; and equity in AbMeta Therapeutics and Adventris Pharmaceuticals. D.J.Z. reports grant/research support from Roche/Genentech. R.J.J. is an employee and stockholder of Roche/Genentech. J.M.T. reports research funding from BMS and Akoya Biosciences; serves as a consultant for BMS, Merck, Astra Zeneca, Genentech, Regeneron, Elephas, Lunaphore, Compugen, and Akoya Biosciences; and holds stock in Akoya Biosciences. The remaining authors declare no competing interests.
Figures













References
-
- Chalabi M et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 26, 566–576 (2020). - PubMed
-
- Cabrita R et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565 (2020). - PubMed
-
- Petitprez F et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 577, 556–560 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA285544/CA/NCI NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- R50 CA243627/CA/NCI NIH HHS/United States
- K08 CA248624/CA/NCI NIH HHS/United States
- R37 CA292056/CA/NCI NIH HHS/United States
- U01CA212007/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- U01 CA212007/CA/NCI NIH HHS/United States
- S10 OD034407/OD/NIH HHS/United States
- U01CA253403/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U01 CA253403/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

