Measurable residual mutated IDH2 before allogeneic transplant for acute myeloid leukemia
- PMID: 39455897
- PMCID: PMC11810785
- DOI: 10.1038/s41409-024-02449-2
Measurable residual mutated IDH2 before allogeneic transplant for acute myeloid leukemia
Abstract
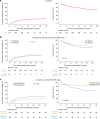
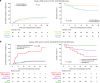
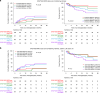
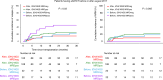
Routine genetic profiling of acute myeloid leukemia (AML) at initial diagnosis has allowed subgroup specific prognostication, drug development, and clinical management strategies. The optimal approach for treatment response assessment for AML subgroups has not yet however been determined. A nationwide cohort of 257 adult patients in first remission (CR1) from AML associated with an IDH2 mutation (IDH2m) undergoing allogeneic transplant during the period 2013-2019 in the United States had rates of relapse and survival three years after transplantation of 24% and 71%, respectively. Pre-transplant clinical flow cytometry assessment was not useful in stratifying patients based on risk of post-transplant relapse or death. DNA-sequencing was performed on CR1 blood collected within 100 days before transplant. Persistent detection of IDH2m was common (51%) and associated with increased relapse and death compared to testing negative. Co-mutation at initial diagnosis with mutated NPM1 and/or FLT3-ITD was common in this cohort (41%) and use of these validated MRD markers provided superior stratification compared to IDH2m testing. Patients testing negative for IDH2m prior to transplant had low relapse-related death, regardless of conditioning intensity. Post-transplant relapse rates for those with persistently detectable IDH2m in pre-transplant remission were lower after the FDA approval of enasidenib in August 2017.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: Protected health information for research was collected and maintained in CIBMTR’s capacity as a public health authority under the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. All patients provided written informed consent for participation in the National Marrow Donor Program institutional review board–approved CIBMTR database (NCT01166009) and repository (NCT04920474) research protocols. Research was performed in compliance with all applicable federal regulations pertaining to the protection of human research participants and with the approval of the CIBMTR observational research group.
Figures


References
-
- Sasaki K, Ravandi F, Kadia TM, DiNardo CD, Short NJ, Borthakur G, et al. De novo acute myeloid leukemia: A population-based study of outcome in the United States based on the Surveillance, Epidemiology, and End Results (SEER) database, 1980 to 2017. Cancer. 2021;127:2049–61. 10.1002/cncr.33458. - PMC - PubMed
-
- Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Gorlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–98. 10.1182/blood-2016-01-693879. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

