Nested patterns of commensals and endosymbionts in microbial communities of mosquito vectors
- PMID: 39455905
- PMCID: PMC11520040
- DOI: 10.1186/s12866-024-03593-x
Nested patterns of commensals and endosymbionts in microbial communities of mosquito vectors
Abstract
Background: Mosquitoes serve as vectors for numerous pathogens, posing significant health risks to humans and animals. Understanding the complex interactions within mosquito microbiota is crucial for deciphering vector-pathogen dynamics and developing effective disease management strategies. Here, we investigated the nested patterns of Wolbachia endosymbionts and Escherichia-Shigella within the microbiota of laboratory-reared Culex pipiens f. molestus and Culex quinquefasciatus mosquitoes. We hypothesized that Wolbachia would exhibit a structured pattern reflective of its co-evolved relationship with both mosquito species, while Escherichia-Shigella would display a more dynamic pattern influenced by environmental factors.
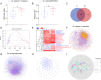
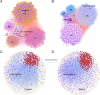
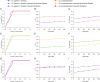
Results: Our analysis revealed different microbial compositions between the two mosquito species, although some microorganisms were common to both. Network analysis revealed distinct community structures and interaction patterns for these bacteria in the microbiota of each mosquito species. Escherichia-Shigella appeared prominently within major network modules in both mosquito species, particularly in module P4 of Cx. pipiens f. molestus, interacting with 93 nodes, and in module Q3 of Cx. quinquefasciatus, interacting with 161 nodes, sharing 55 nodes across both species. On the other hand, Wolbachia appeared in disparate modules: module P3 in Cx. pipiens f. molestus and a distinct module with a single additional taxon in Cx. quinquefasciatus, showing species-specific interactions and no shared taxa. Through computer simulations, we evaluated how the removal of Wolbachia or Escherichia-Shigella affects network robustness. In Cx. pipiens f. molestus, removal of Wolbachia led to a decrease in network connectivity, while Escherichia-Shigella removal had a minimal impact. Conversely, in Cx. quinquefasciatus, removal of Escherichia-Shigella resulted in decreased network stability, whereas Wolbachia removal had minimal effect.
Conclusions: Contrary to our hypothesis, the findings indicate that Wolbachia displays a more dynamic pattern of associations within the microbiota of Culex pipiens f. molestus and Culex quinquefasciatus mosquitoes, than Escherichia-Shigella. The differential effects on network robustness upon Wolbachia or Escherichia-Shigella removal suggest that these bacteria play distinct roles in maintaining community stability within the microbiota of the two mosquito species.
Keywords: Escherichia-Shigella; Wolbachia; Community assembly; Mosquitoes; Nestedness theory.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

