Identification of the optimal reference genes for atrial fibrillation model established by iPSC-derived atrial myocytes
- PMID: 39455925
- PMCID: PMC11515253
- DOI: 10.1186/s12864-024-10922-x
Identification of the optimal reference genes for atrial fibrillation model established by iPSC-derived atrial myocytes
Abstract
Background: Atrial fibrillation (AF) stands as a prevalent and detrimental arrhythmic disorder, characterized by intricate pathophysiological mechanisms. The availability of reliable and reproducible AF models is pivotal in unraveling the underlying mechanisms of this complex condition. Unfortunately, the researchers are still confronted with the absence of consistent in vitro AF models, hindering progress in this crucial area of research.
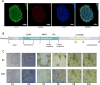
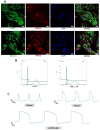
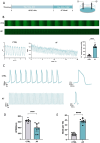
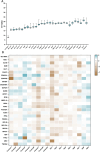
Methods: Human induced pluripotent stem cells derived atrial myocytes (hiPSC-AMs) were generated based on the GiWi methods and were verified by whole-cell patch clamp, immunofluorescent staining, and flow cytometry. Then hiPSC-AMs were employed to establish the AF model by HS. Whole-cell patch clamp technique and calcium imaging were used to identify the AF model. The stability of 29 reference genes was evaluated using delta-Ct, GeNorm, NormFinder, and BestKeeper algorithms; RESULTS: HiPSC-AMs displayed atrial myocyte action potentials and expressed the atrial-specific protein MLC-2 A and NR2F2, about 70% of the cardiomyocytes were MLC-2 A positive. After HS, hiPSC-AMs showed a significant increase in beating frequency, a shortened action potential duration, and increased calcium transient frequency. Of the 29 candidate genes, the top five most stably ranked genes were ABL1, RPL37A, POP4, RPL30, and EIF2B1. After normalization using ABL1, KCNJ2 was significantly upregulated in the AF model; Conclusions: In the hiPSC-AMs AF model established by HS, ABL1 provides greater normalization efficiency than commonly used GAPDH.
Keywords: Atrial fibrillation; Atrial fibrillation model; Electrical stimulation; Induced pluripotent stem cells; Quantitative real-time polymerase chain reaction; Reference gene.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Grants and funding
- 2022YFS0610/Sichuan Province Science and Technology Support Program
- 2022YFS0617/Sichuan Province Science and Technology Support Program
- 2021LZXNYD-J26/Luzhou Science and Technology Bureau
- 2021LZXNYD-Z07/Luzhou Science and Technology Bureau
- KeyME- 2023-01/Key Laboratory of Medical Electrophysiology of Ministry of Education
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

