Identification of molecular subtypes based on bile acid metabolism in cholangiocarcinoma
- PMID: 39455933
- PMCID: PMC11515294
- DOI: 10.1186/s12885-024-13081-0
Identification of molecular subtypes based on bile acid metabolism in cholangiocarcinoma
Abstract
Background: Cholangiocarcinoma is a highly heterogeneous tumor with bile acid metabolism involving in its development. The aim of this study was to characterize bile acid metabolism and identify specific subtypes to better stratify cholangiocarcinoma patients for individualized treatment and prognostic assessment.
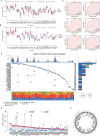
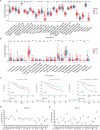
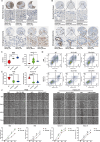
Methods: A total of 30 bile acids were quantified using the ultra-performance liquid chromatography tandem mass spectrometry. Using Consensus clustering, the molecular subtypes related to bile acid metabolism were identified. The prognosis, clinicopathologic characteristics, immune landscape, and therapeutic response were compared between these subtypes. The single-cell RNA sequencing (scRNA-seq) analysis and preliminary cell experiment were also conducted to verify our findings.
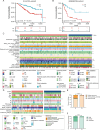
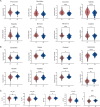
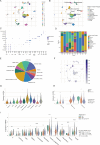
Results: The altered bile acid profile and genetic variation of bile acid metabolism-related genes in cholangiocarcinoma were demonstrated. The cholangiocarcinoma was categorized into bile acid metabolism-active and -inactive subtypes with different prognoses, clinicopathologic characteristics, tumor microenvironments (TME) and therapeutic responses. This categorization was reproducible and predictable. Specifically, the bile acid metabolism-active subtype showed a poor prognosis with an immunosuppressive microenvironment and an inactive response to immunotherapy, while the bile acid metabolism-inactive subtype showed the opposite characteristics. Moreover, the scRNA-seq revealed that immunotherapy altered bile acid metabolism in TME of cholangiocarcinoma. Finally, a prognostic signature related to bile acid metabolism was developed, which exhibited strong power for prognostic assessment of cholangiocarcinoma. Consistently, these results were verified by immunohistochemistry, cell proliferation, migration, and apoptosis assays.
Conclusion: In conclusion, a novel cholangiocarcinoma classification based on bile acid metabolism was established. This classification was significant for the estimation of TME and prognosis.
Keywords: Bile acids; Cholangiocarcinoma; Prognosis; Subtypes; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

