Repetitive transcranial magnetic stimulation alleviates motor impairment in Parkinson's disease: association with peripheral inflammatory regulatory T-cells and SYT6
- PMID: 39456006
- PMCID: PMC11515224
- DOI: 10.1186/s13024-024-00770-4
Repetitive transcranial magnetic stimulation alleviates motor impairment in Parkinson's disease: association with peripheral inflammatory regulatory T-cells and SYT6
Abstract
Background: Repetitive transcranial magnetic stimulation (rTMS) has been used to treat various neurological disorders. However, the molecular mechanism underlying the therapeutic effect of rTMS on Parkinson's disease (PD) has not been fully elucidated. Neuroinflammation like regulatory T-cells (Tregs) appears to be a key modulator of disease progression in PD. If rTMS affects the peripheral Tregs in PD remains unknown.
Methods: Here, we conducted a prospective clinical study (Chinese ClinicalTrials. gov: ChiCTR 2100051140) involving 54 PD patients who received 10-day rTMS (10 Hz) stimulation on the primary motor cortex (M1) region or sham treatment. Clinical and function assessment as well as flow cytology study were undertaken in 54 PD patients who were consecutively recruited from the department of neurology at Zhujiang hospital between September 2021 and January 2022. Subsequently, we implemented flow cytometry analysis to examine the Tregs population in spleen of MPTP-induced PD mice that received rTMS or sham treatment, along with quantitative proteomic approach reveal novel molecular targets for Parkinson's disease, and finally, the RNA interference method verifies the role of these new molecular targets in the treatment of PD.
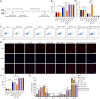
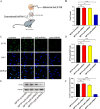
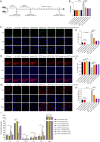
Results: We demonstrated that a 10-day rTMS treatment on the M1 motor cortex significantly improved motor dysfunction in PD patients. The beneficial effects persisted for up to 40 days, and were associated with an increase in peripheral Tregs. There was a positive correlation between Tregs and motor improvements in PD cases. Similarly, a 10-day rTMS treatment on the brains of MPTP-induced PD mice significantly ameliorated motor symptoms. rTMS reversed the downregulation of circulating Tregs and tyrosine hydroxylase neurons in these mice. It also increased anti-inflammatory mediators, deactivated microglia, and decreased inflammatory cytokines. These effects were blocked by administration of a Treg inhibitor anti-CD25 antibody in MPTP-induced PD mice. Quantitative proteomic analysis identified TLR4, TH, Slc6a3 and especially Syt6 as the hub node proteins related to Tregs and rTMS therapy. Lastly, we validated the role of Treg and rTMS-related protein syt6 in MPTP mice using the virus interference method.
Conclusions: Our clinical and experimental studies suggest that rTMS improves motor function by modulating the function of Tregs and suppressing toxic neuroinflammation. Hub node proteins (especially Syt6) may be potential therapeutic targets.
Trial registration: Chinese ClinicalTrials, ChiCTR2100051140. Registered 15 December 2021, https://www.chictr.org.cn/bin/project/edit?pid=133691.
Keywords: Motor dysfunction, syt6; Neuro-inflammation; Parkinson’s disease; Regulatory T cells; Repetitive transcranial magnetic stimulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Tan EK, Chao YX, West A, Chan LL, Poewe W, Jankovic J. Parkinson disease and the immune system - associations, mechanisms and therapeutics. Nat Rev Neurol. 2020;16(6):303–18. - PubMed
-
- Hirsch EC, Hunot S. Neuroinflammation in Parkinson’s disease: a target for neuroprotection? Lancet Neurol. 2009;8(4):382–97. - PubMed
-
- Yang W, Chang Z, Que R, Weng G, Deng B, Wang T, Huang Z, Xie F, Wei X, Yang Q, et al. Contra-Directional Expression of Plasma Superoxide Dismutase with Lipoprotein Cholesterol and High-Sensitivity C-reactive Protein as Important Markers of Parkinson’s Disease Severity. Front Aging Neurosci. 2020;12:53. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

