RNA-seq validation of microRNA expression signatures for precision melanoma diagnosis and prognostic stratification
- PMID: 39456086
- PMCID: PMC11515382
- DOI: 10.1186/s12920-024-02028-w
RNA-seq validation of microRNA expression signatures for precision melanoma diagnosis and prognostic stratification
Abstract
Background: New diagnostic tools are needed to improve the diagnosis and risk stratification of cutaneous melanoma. Disease-specific microRNA signatures have been previously described via NanoString profiling of solid biopsy tissue and plasma. This study validated these signatures via next-generation sequencing technology and compared their performance against clinical metrics and other published melanoma signatures.
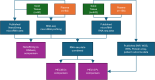
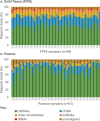
Methods: RNA from 64 plasma and 60 FFPE biopsy samples from individuals with invasive melanoma or related benign/control phenotypes was extracted and enriched for microRNA. RNA sequencing was performed to compute MEL38/MEL12 signature scores. The results were evaluated with published NanoString and RNA sequencing datasets, comprising 548 solid tissue samples and 217 plasma samples, to predict disease status and patient outcome.
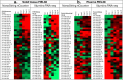
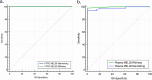
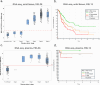
Results: The MEL38 diagnostic signature classifies patients into discrete diagnostic groups via RNA sequencing in either solid tissue or plasma (P < 0.001). In solid tissue, the prognostic MEL12 signature stratifies patients into low-, intermediate- and high-risk groups, independent of clinical covariates. The hazard ratios for 10-year overall survival, based on observed survival intervals, were 2.2 (MEL12 high-risk vs low-risk, P < 0.001) and 1.8 (intermediate-risk vs low-risk, P < 0.001), outperforming other published prognostic models. MEL12 also exhibited prognostic significance in the plasma of 42 patients with invasive disease.
Conclusions: The MEL38 and MEL12 signatures can be assessed in either solid tissue or plasma using RNA-seq and are strong predictors of disease state and patient outcome. Compared with other genomic methods, MEL12 was shown to be the strongest predictor of poor prognosis. MicroRNA expression profiling offers objective, accurate genomic information about a patient's likelihood of invasive melanoma and prognosis.
Keywords: Cancer; Gene expression; Melanoma; MicroRNA signatures; Precision medicine; RNA-seq.
© 2024. The Author(s).
Conflict of interest statement
CGL and RVL are employed and have equity in Geneseq Biosciences. LC is an employee of Australian Clinical Labs.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

