Integration of fungal transcriptomics and metabolomics provides insights into the early interaction between the ORM fungus Tulasnella sp. and the orchid Serapias vomeracea seeds
- PMID: 39456087
- PMCID: PMC11503967
- DOI: 10.1186/s43008-024-00165-6
Integration of fungal transcriptomics and metabolomics provides insights into the early interaction between the ORM fungus Tulasnella sp. and the orchid Serapias vomeracea seeds
Abstract
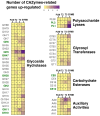
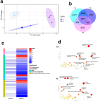
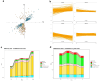
In nature, germination of orchid seeds and early plant development rely on a symbiotic association with orchid mycorrhizal (ORM) fungi. These fungi provide the host with the necessary nutrients and facilitate the transition from embryos to protocorms. Despite recent advances in omics technologies, our understanding of this symbiosis remains limited, particularly during the initial stages of the interaction. To address this gap, we employed transcriptomics and metabolomics to investigate the early responses occurring in the mycorrhizal fungus Tulasnella sp. isolate SV6 when co-cultivated with orchid seeds of Serapias vomeracea. The integration of data from gene expression and metabolite profiling revealed the activation of some fungal signalling pathways before the establishment of the symbiosis. Prior to seed contact, an indole-related metabolite was produced by the fungus, and significant changes in the fungal lipid profile occurred throughout the symbiotic process. Additionally, the expression of plant cell wall-degrading enzymes (PCWDEs) was observed during the pre-symbiotic stage, as the fungus approached the seeds, along with changes in amino acid metabolism. Thus, the dual-omics approach employed in this study yielded novel insights into the symbiotic relationship between orchids and ORM fungi and suggest that the ORM fungus responds to the presence of the orchid seeds prior to contact.
Keywords: CAZymes; Mycorrhizal fungi; Omics; Orchids; Symbiosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Atanasova L, Dubey M, Grujić M, Gudmundsson M, Lorenz C, Sandgren M, Kubicek CP, Jensen DF, Karlsson M (2018) Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbiol. 10.1186/s12866-018-1310-9 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
