A Novel Peptide from VP1 of EV-D68 Exhibits Broad-Spectrum Antiviral Activity Against Human Enteroviruses
- PMID: 39456264
- PMCID: PMC11506774
- DOI: 10.3390/biom14101331
A Novel Peptide from VP1 of EV-D68 Exhibits Broad-Spectrum Antiviral Activity Against Human Enteroviruses
Abstract
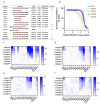
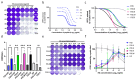
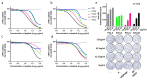
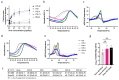
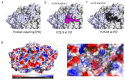
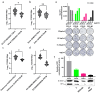
Enteroviruses have been a historical concern since the identification of polioviruses in humans. Wild polioviruses have almost been eliminated, while multiple species of non-polio enteroviruses and their variants co-circulate annually. To date, at least 116 types have been found in humans and are grouped into the species Enterovirus A-D and Rhinovirus A-C. However, there are few available antiviral drugs, especially with a universal pharmaceutical effect. Here, we demonstrate that peptide P25 from EV-D68 has broad antiviral activity against EV A-D enteroviruses in vitro. P25, derived from the HI loop and β-I sheet of VP1, operates through a conserved hydrophilic motif -R---K-K--K- and the hydrophobic F near the N-terminus. It could prevent viral infection of EV-A71 by competing for the heparan sulfate (HS) receptor, binding and stabilizing virions by suppressing the release of the viral genome. P25 also inhibited the generation of infectious viral particles by reducing viral protein synthesis. The molecular docking revealed that P25 might bind to the pocket opening area, a potential target for broad-spectrum antivirals. Our findings implicate the multiple antiviral effects of peptide P25, including blocking viral binding to the HS receptor, impeding viral genome release, and reducing progeny particles, which could be a novel universal anti-enterovirus drug candidate.
Keywords: EV-D68; antiviral; broad-spectrum; human enteroviruses; peptide.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- World Health Organization Poliomyelitis. [(accessed on 11 May 2024)]. Available online: https://www.who.int/news-room/fact-sheets/detail/poliomyelitis.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

