Identification of Multifunctional Putative Bioactive Peptides in the Insect Model Red Palm Weevil (Rhynchophorus ferrugineus)
- PMID: 39456265
- PMCID: PMC11506011
- DOI: 10.3390/biom14101332
Identification of Multifunctional Putative Bioactive Peptides in the Insect Model Red Palm Weevil (Rhynchophorus ferrugineus)
Abstract
Innate immunity, the body's initial defense against bacteria, fungi, and viruses, heavily depends on antimicrobial peptides (AMPs), which are small molecules produced by all living organisms. Insects, with their vast biodiversity, are one of the most abundant and innovative sources of AMPs. In this study, AMPs from the red palm weevil (RPW) Rhynchophorus ferrugineus (Coleoptera: Curculionidae), a known invasive pest of palm species, were examined. The AMPs were identified in the transcriptomes from different body parts of male and female adults, under different experimental conditions, including specimens collected from the field and those reared in the laboratory. The RPW transcriptomes were examined to predict antimicrobial activity, and all sequences putatively encoding AMPs were analyzed using several machine learning algorithms available in the CAMPR3 database. Additionally, anticancer, antiviral, and antifungal activity of the peptides were predicted using iACP, AVPpred, and Antifp server tools, respectively. Physicochemical parameters were assessed using the Antimicrobial Peptide Database Calculator and Predictor. From these analyses, 198 putatively active peptides were identified, which can be tested in future studies to validate the in silico predictions. Genome-wide analysis revealed that several AMPs have predominantly emerged through gene duplication. Noticeably, we detect a newly originated defensin allele from an ancestral defensin via the deletion of two amino acids following gene duplication in RPW, which may confer an enhanced resilience to microbial infection. Our study shed light on AMP gene families and shows that high duplication and deletion rates are essential to achieve a diversity of antimicrobial mechanisms; hence, we propose the RPW AMPs as a model for exploring gene duplication and functional variations against microbial infection.
Keywords: ACPs; AFPs; AVPs; antimicrobial peptides; bioinformatic tools; gene duplication; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest. P.F., B.A., and A.P. may have a financial interest in RPW AMP(s) patent applications related to drug development.
Figures
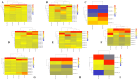



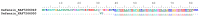
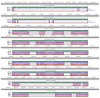
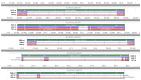
References
-
- Moretta A., Scieuzo C., Petrone A.M., Salvia R., Manniello M.D., Franco A., Lucchetti D., Vassallo A., Vogel H., Sgambato A., et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021;11:668632. doi: 10.3389/fcimb.2021.668632. - DOI - PMC - PubMed
-
- Brandenburg L.O., Merres J., Albrecht L.J., Varoga D., Pufe T. Antimicrobial peptides: Multifunctional drugs for different applications. Polymers. 2012;4:539–560. doi: 10.3390/polym4010539. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

