Functional Characterization of the Ciliate Stylonychia lemnae Serotonin N-Acetyltransferase, a Pivotal Enzyme in Melatonin Biosynthesis and Its Overexpression Leads to Peroxidizing Herbicide Tolerance in Rice
- PMID: 39456431
- PMCID: PMC11505474
- DOI: 10.3390/antiox13101177
Functional Characterization of the Ciliate Stylonychia lemnae Serotonin N-Acetyltransferase, a Pivotal Enzyme in Melatonin Biosynthesis and Its Overexpression Leads to Peroxidizing Herbicide Tolerance in Rice
Abstract
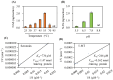
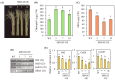
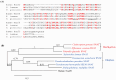
Serotonin N-acetyltransferase (SNAT) is a pivotal enzyme for melatonin biosynthesis in all living organisms. It catalyzes the conversion of serotonin to N-acetylserotonin (NAS) or 5-methoxytrypytamine (5-MT) to melatonin. In contrast to animal- and plant-specific SNAT genes, a novel clade of archaeal SNAT genes has recently been reported. In this study, we identified homologues of archaeal SNAT genes in ciliates and dinoflagellates, but no animal- or plant-specific SNAT homologues. Archaeal SNAT homologue from the ciliate Stylonychia lemnae was annotated as a putative N-acetyltransferase. To determine whether the putative S. lemnae SNAT (SlSNAT) exhibits SNAT enzyme activity, we chemically synthesized and expressed the full-length SlSNAT coding sequence (CDS) in Escherichia coli, from which the recombinant SlSNAT protein was purified by Ni2+ affinity column chromatography. The recombinant SlSNAT exhibited SNAT enzyme activity toward serotonin (Km = 776 µM) and 5-MT (Km = 246 µM) as substrates. Furthermore, SlSNAT-overexpressing (SlSNAT-OE) transgenic rice plants showed higher levels of melatonin synthesis than wild-type controls. The SlSNAT-OE rice plants exhibited delayed leaf senescence and tolerance against treatment with the reactive oxygen species (ROS)-inducing herbicide butafenacil by decreasing hydrogen peroxide (H2O2) and malondialdehyde (MDA) levels, suggesting that melatonin alleviates ROS production in vivo.
Keywords: Osl20; Stylonichia lemnae; archaea; ciliophoran; melatonin; serotonin N-acetyltransferase; transgenic rice.
Conflict of interest statement
The authors declared no conflicts of interest.
Figures




References
-
- Hardeland R. Melatonin in the evolution of plants and other phototrophs. Melatonin Res. 2019;2:10–36. doi: 10.32794/mr11250029. - DOI
-
- Lerner A.B., Case J.D., Takahashi Y. Isolation of melatonin, a pineal factor that lightness melanocytes. J. Am. Chem. Soc. 1958;80:2587. doi: 10.1021/ja01543a060. - DOI
-
- Lerner A.B., Case J.D., Heinzelmann R.V. Structure of melatonin. J. Am. Chem. Soc. 1959;81:6084–6085. doi: 10.1021/ja01531a060. - DOI
-
- Suzen S., Atayik M.C., Sirinzade H., Entezari B., Gurer-Orhan H., Cakatay U. Melatonin and redox homeostasis. Melatonin Res. 2022;5:304–324. doi: 10.32794/mr112500134. - DOI
LinkOut - more resources
Full Text Sources

