A Single-Cell Atlas of the Substantia Nigra Reveals Therapeutic Effects of Icaritin in a Rat Model of Parkinson's Disease
- PMID: 39456437
- PMCID: PMC11505506
- DOI: 10.3390/antiox13101183
A Single-Cell Atlas of the Substantia Nigra Reveals Therapeutic Effects of Icaritin in a Rat Model of Parkinson's Disease
Abstract
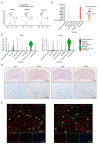
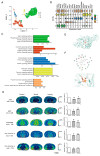
Degeneration and death of dopaminergic neurons in the substantia nigra of the midbrain are the main pathological changes in Parkinson's disease (PD); however, the mechanism underlying the selective vulnerability of specific neuronal populations in PD remains unclear. Here, we used single-cell RNA sequencing to identify seven cell clusters, including oligodendrocytes, neurons, astrocytes, oligodendrocyte progenitor cells, microglia, synapse-rich cells (SRCs), and endothelial cells, in the substantia nigra of a rotenone-induced rat model of PD based on marker genes and functional definitions. We found that SRCs were a previously unidentified cell subtype, and the tight interactions between SRCs and other cell populations can be improved by icaritin, which is a flavonoid extracted from Epimedium sagittatum Maxim. and exerts anti-neuroinflammatory, antioxidant, and immune-improving effects in PD. We also demonstrated that icaritin bound with transcription factors of SRCs, and icaritin application modulated synaptic characterization of SRCs, neuroinflammation, oxidative stress, and survival of dopaminergic neurons, and improved abnormal energy metabolism, amino acid metabolism, and phospholipase D metabolism of astrocytes in the substantia nigra of rats with PD. Moreover, icaritin supplementation also promotes the recovery of the physiological homeostasis of the other cell clusters to delay the pathogenesis of PD. These data uncovered previously unknown cellular diversity in a rat model of Parkinson's disease and provide insights into the promising therapeutic potential of icaritin in PD.
Keywords: Parkinson’s disease; icaritin; neuroinflammation; oxidative stress; rotenone; single-cell RNA sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources

