Functional Assessments of Gynecologic Cancer Models Highlight Differences Between Single-Node Inhibitors of the PI3K/AKT/mTOR Pathway and a Pan-PI3K/mTOR Inhibitor, Gedatolisib
- PMID: 39456616
- PMCID: PMC11505998
- DOI: 10.3390/cancers16203520
Functional Assessments of Gynecologic Cancer Models Highlight Differences Between Single-Node Inhibitors of the PI3K/AKT/mTOR Pathway and a Pan-PI3K/mTOR Inhibitor, Gedatolisib
Abstract
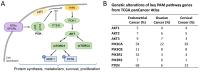
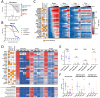
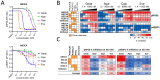
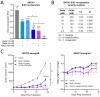
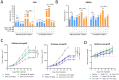
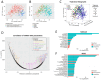
Background/Objectives: The PI3K/AKT/mTOR (PAM) pathway is frequently activated in gynecological cancers. Many PAM inhibitors selectively target single PAM pathway nodes, which can lead to reduced efficacy and increased drug resistance. To address these limitations, multiple PAM pathway nodes may need to be inhibited. Gedatolisib, a well-tolerated panPI3K/mTOR inhibitor targeting all Class I PI3K isoforms, mTORC1 and mTORC2, could represent an effective treatment option for patients with gynecologic cancers. Methods: Gedatolisib and other PAM inhibitors (e.g., alpelisib, capivasertib, and everolimus) were tested in endometrial, ovarian, and cervical cancer cell lines by using cell viability, cell proliferation, and flow cytometry assays. Xenograft studies evaluated gedatolisib in combination with a CDK4/6 inhibitor (palbociclib) or an anti-estrogen (fulvestrant). A pseudo-temporal transcriptomic trajectory of endometrial cancer clinical progression was computationally modeled employing data from 554 patients to correlate non-clinical studies with a potential patient group. Results: Gedatolisib induced a substantial decrease in PAM pathway activity in association with the inhibition of cell cycle progression and the decreased cell viability in vitro. Compared to single-node PAM inhibitors, gedatolisib exhibited greater growth-inhibitory effects in almost all cell lines, regardless of the PAM pathway mutations. Gedatolisib combined with either fulvestrant or palbociclib inhibited tumor growth in endometrial and ovarian cancer xenograft models. Conclusions: Gedatolisib in combination with other therapies has shown an acceptable safety profile and promising preliminary efficacy in clinical studies with various solid tumor types. The non-clinical data presented here support the development of gedatolisib combined with CDK4/6 inhibitors and/or hormonal therapy for gynecologic cancer treatment.
Keywords: PI3K/AKT/mTOR pathway; endometrial cancer; gedatolisib; ovarian cancer.
Conflict of interest statement
S.R., A.B., A.S., I.M., J.M. and L.L. are all employed by and/or have ownership interest in Celcuity, Inc. A.S.M. performed his work as a temporary intern at Celcuity, Inc.
Figures

Similar articles
-
Functional Analysis of the PI3K/AKT/mTOR Pathway Inhibitor, Gedatolisib, Plus Fulvestrant with and Without Palbociclib in Breast Cancer Models.Int J Mol Sci. 2025 Jun 18;26(12):5844. doi: 10.3390/ijms26125844. Int J Mol Sci. 2025. PMID: 40565304 Free PMC article.
-
Assessments of prostate cancer cell functions highlight differences between a pan-PI3K/mTOR inhibitor, gedatolisib, and single-node inhibitors of the PI3K/AKT/mTOR pathway.Mol Oncol. 2025 Jan;19(1):225-247. doi: 10.1002/1878-0261.13703. Epub 2024 Aug 2. Mol Oncol. 2025. PMID: 39092562 Free PMC article.
-
Gedatolisib shows superior potency and efficacy versus single-node PI3K/AKT/mTOR inhibitors in breast cancer models.NPJ Breast Cancer. 2024 Jun 5;10(1):40. doi: 10.1038/s41523-024-00648-0. NPJ Breast Cancer. 2024. PMID: 38839777 Free PMC article.
-
mTOR inhibitors and their clinical application in cervical, endometrial and ovarian cancers: a critical review.Gynecol Oncol. 2014 May;133(2):375-81. doi: 10.1016/j.ygyno.2014.02.017. Epub 2014 Feb 18. Gynecol Oncol. 2014. PMID: 24556063 Review.
-
Drugging the PI3K/AKT/mTOR Pathway in ER+ Breast Cancer.Int J Mol Sci. 2023 Feb 24;24(5):4522. doi: 10.3390/ijms24054522. Int J Mol Sci. 2023. PMID: 36901954 Free PMC article. Review.
Cited by
-
Functional Analysis of the PI3K/AKT/mTOR Pathway Inhibitor, Gedatolisib, Plus Fulvestrant with and Without Palbociclib in Breast Cancer Models.Int J Mol Sci. 2025 Jun 18;26(12):5844. doi: 10.3390/ijms26125844. Int J Mol Sci. 2025. PMID: 40565304 Free PMC article.
-
Molecular pathways in reproductive cancers: a focus on prostate and ovarian cancer.Cancer Cell Int. 2025 Feb 3;25(1):33. doi: 10.1186/s12935-025-03658-5. Cancer Cell Int. 2025. PMID: 39901204 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

