The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas
- PMID: 39456618
- PMCID: PMC11505613
- DOI: 10.3390/cancers16203524
The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas
Abstract
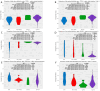
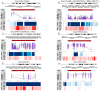
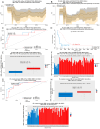
Background: Changes in DNA methylation patterns are a pivotal mechanism of carcinogenesis. In some tumors, aberrant methylation precedes genetic changes, while gene expression may be more frequently modified due to methylation alterations than by mutations. Methods: Herein, 128 serous ovarian tumors were analyzed, including borderline ovarian tumors (BOTS) with (BOT.V600E) and without (BOT) the BRAF V600E mutation, low-grade (lg), and high-grade (hg) ovarian cancers (OvCa). The methylome of the samples was profiled with Infinium MethylationEPIC microarrays. Results: The biggest number of differentially methylated (DM) CpGs and regions (DMRs) was found between lgOvCa and hgOvCa. By contrast, the BOT.V600E tumors had the lowest number of DM CpGs and DMRs compared to all other groups and, in relation to BOT, their genome was strongly downmethylated. Remarkably, the ten most significant DMRs, discriminating BOT from lgOvCa, encompassed the MHC region on chromosome 6. We also identified hundreds of DMRs, being of potential use as predictive biomarkers in BOTS and hgOvCa. DMRs with the best discriminative capabilities overlapped the following genes: BAIAP3, IL34, WNT10A, NEU1, SLC44A4, and HMOX1, TCN2, PES1, RP1-56J10.8, ABR, NCAM1, RP11-629G13.1, AC006372.4, NPTXR in BOTS and hgOvCa, respectively. Conclusions: The global genome-wide hypomethylation positively correlates with the increasing aggressiveness of ovarian tumors. We also assume that the immune system may play a pivotal role in the transition from BOTS to lgOvCa. Given that the BOT.V600E tumors had the lowest number of DM CpGs and DMRs compared to all other groups, when methylome is considered, such tumors might be placed in-between BOT and OvCa.
Keywords: DNA methylation; biomarkers; methylation microarrays; serous borderline ovarian tumor; serous ovarian carcinoma.
Conflict of interest statement
The authors declare no conflicts of interest. The sponsors had no role in the design, execution, interpretation, or writing of the study.
Figures


References
-
- Bourdel N., Huchon C., Abdel Wahab C., Azaïs H., Bendifallah S., Bolze P.-A., Brun J.-L., Canlorbe G., Chauvet P., Chereau E., et al. Borderline Ovarian Tumors: French Guidelines from the CNGOF. Part 2. Surgical Management, Follow-up, Hormone Replacement Therapy, Fertility Management and Preservation. J. Gynecol. Obstet. Hum. Reprod. 2021;50:101966. doi: 10.1016/j.jogoh.2020.101966. - DOI - PubMed
-
- Timor-Tritsch I.E., Foley C.E., Brandon C., Yoon E., Ciaffarrano J., Monteagudo A., Mittal K., Boyd L. New Sonographic Marker of Borderline Ovarian Tumor: Microcystic Pattern of Papillae and Solid Components. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2019;54:395–402. doi: 10.1002/uog.20283. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

