Effects of Differentially Methylated CpG Sites in Enhancer and Promoter Regions on the Chromatin Structures of Target LncRNAs in Breast Cancer
- PMID: 39456830
- PMCID: PMC11507307
- DOI: 10.3390/ijms252011048
Effects of Differentially Methylated CpG Sites in Enhancer and Promoter Regions on the Chromatin Structures of Target LncRNAs in Breast Cancer
Abstract
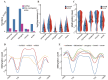
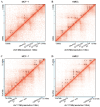
Aberrant DNA methylation plays a crucial role in breast cancer progression by regulating gene expression. However, the regulatory pattern of DNA methylation in long noncoding RNAs (lncRNAs) for breast cancer remains unclear. In this study, we integrated gene expression, DNA methylation, and clinical data from breast cancer patients included in The Cancer Genome Atlas (TCGA) database. We examined DNA methylation distribution across various lncRNA categories, revealing distinct methylation characteristics. Through genome-wide correlation analysis, we identified the CpG sites located in lncRNAs and the distally associated CpG sites of lncRNAs. Functional genome enrichment analysis, conducted through the integration of ENCODE ChIP-seq data, revealed that differentially methylated CpG sites (DMCs) in lncRNAs were mostly located in promoter regions, while distally associated DMCs primarily acted on enhancer regions. By integrating Hi-C data, we found that DMCs in enhancer and promoter regions were closely associated with the changes in three-dimensional chromatin structures by affecting the formation of enhancer-promoter loops. Furthermore, through Cox regression analysis and three machine learning models, we identified 11 key methylation-driven lncRNAs (DIO3OS, ELOVL2-AS1, MIAT, LINC00536, C9orf163, AC105398.1, LINC02178, MILIP, HID1-AS1, KCNH1-IT1, and TMEM220-AS1) that were associated with the survival of breast cancer patients and constructed a prognostic risk scoring model, which demonstrated strong prognostic performance. These findings enhance our understanding of DNA methylation's role in lncRNA regulation in breast cancer and provide potential biomarkers for diagnosis.
Keywords: DNA methylation; Hi-C; breast cancer; enhancer; lncRNA; promoter.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Chhikara B.S., Parang K. Global Cancer Statistics 2022: The Trends Projection Analysis. Chem. Biol. Lett. 2023;10:451.
-
- Rodríguez Bautista R., Ortega Gómez A., Hidalgo Miranda A., Zentella Dehesa A., Villarreal-Garza C., Ávila-Moreno F., Arrieta O. Long Non-Coding RNAs: Implications in Targeted Diagnoses, Prognosis, and Improved Therapeutic Strategies in Human Non- and Triple-Negative Breast Cancer. Clin. Epigenet. 2018;10:88. doi: 10.1186/s13148-018-0514-z. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

