Contrast Relative Humidity Response of Diverse Cowpea (Vigna unguiculata (L.) Walp.) Genotypes: Deep Study Using RNAseq Approach
- PMID: 39456837
- PMCID: PMC11507454
- DOI: 10.3390/ijms252011056
Contrast Relative Humidity Response of Diverse Cowpea (Vigna unguiculata (L.) Walp.) Genotypes: Deep Study Using RNAseq Approach
Abstract
Cowpea (Vigna unguiculata (L.) Walp.) is appreciated for its suitability for cultivation and obtaining good yields in relatively extreme farming conditions. It is resistant to high temperatures and drought. Moreover, food products prepared from Vigna are rich in many nutrients such as proteins, amino acids, carbohydrates, minerals, fiber, vitamins, and other bioactive compounds. However, in East and Southeast Asia, where the products of this crop are in demand, the climate is characterized by excessive humidity. Under these conditions, the vast majority of cowpea varieties tend to have indeterminate growth (elongated shoot length) and are unsuitable for mechanized harvesting. The molecular mechanisms for tolerance to high relative humidity remain the least studied in comparison with those for other abiotic stress factors (drought, heat, cold, flooding, etc.). The purpose of the work was to reveal and investigate differentially expressed genes in cowpea accessions having contrasting growth habits (determinate and indeterminate) under humid and drought conditions. We performed RNA-seq analysis using selected cowpea accessions from the VIR collection. Among the genotypes used, some have significant changes in their plant architecture in response to high relative humidity, while others were tolerant to these conditions. In total, we detected 1697 upregulated and 1933 downregulated genes. The results showed that phytohormone-related genes are involved in cowpea response to high relative humidity. DEGs associated with jasmonic acid signaling are proposed to be key contributors in the maintenance of compact architecture under humid conditions.
Keywords: cowpea; differentially expressed gene; high relative humidity; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

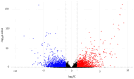

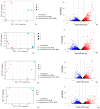





References
-
- Timko M.P., Ehlers J.D., Roberts P.A. Cowpea. In: Kole C., editor. Genome Mapping and Molecular Breeding in Plants. Pulses, Sugar and Tuber Crops. Volume 3. Springer; Berlin/Heidelberg, Germany: 2007. pp. 49–67.
-
- Gerrano A.S., Adebola P.O., van Rensburg W.S.J., Venter S.L. Genetic Variability and Heritability Estimates of Nutritional Composition in the Leaves of Selected Cowpea Genotypes [Vigna unguiculata (L.) Walp] HortScience. 2015;50:1435–1440. doi: 10.21273/HORTSCI.50.10.1435. - DOI
-
- Gerrano A.S., van Rensburg W.S.J., Venter S.L., Shargie N.G., Amelework B.A., Shimelis H.A., Labuschagne M.T. Selection of Cowpea Genotypes Based on Grain Mineral and Total Protein Content. Acta Agric. Scand. Sect. B Soil Plant Sci. 2019;69:155–166. doi: 10.1080/09064710.2018.1520290. - DOI
-
- Gerrano A.S., van Rensburg W.S.J., Adebola P.O. Nutritional Composition of Immature Pods in Selected Cowpea [Vigna unguiculata (L.) Walp.] Genotypes in South Africa. Aust. J. Crop Sci. 2017;11:134–141. doi: 10.21475/ajcs.17.11.02.p72. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

