Single-Base Methylome Analysis of Sweet Cherry (Prunus avium L.) on Dwarfing Rootstocks Reveals Epigenomic Differences Associated with Scion Dwarfing Conferred by Grafting
- PMID: 39456883
- PMCID: PMC11508414
- DOI: 10.3390/ijms252011100
Single-Base Methylome Analysis of Sweet Cherry (Prunus avium L.) on Dwarfing Rootstocks Reveals Epigenomic Differences Associated with Scion Dwarfing Conferred by Grafting
Abstract
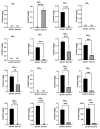
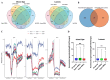
Plant grafting using dwarfing rootstocks is one of the important cultivation measures in the sweet cherry (Prunus avium) industry. In this work, we aimed to explore the effects of the dwarfing rootstock "Pd1" (Prunus tomentosa) on sweet cherry 'Shuguang2' scions by performing morphological observations using the paraffin slice technique, detecting GA (gibberellin) and IAA (auxin) contents using UPLC-QTRAP-MS (ultra-performance liquid chromatography coupled with a hybrid triple quadrupole-linear ion trap mass spectrometer), and implementing integration analyses of the epigenome and transcriptome using whole-genome bisulfite sequencing and transcriptome sequencing. Anatomical analysis indicated that the cell division ability of the SAM (shoot apical meristem) in dwarfing plants was reduced. Pd1 rootstock significantly decreased the levels of GAs and IAA in sweet cherry scions. Methylome analysis showed that the sweet cherry genome presented 15.2~18.6%, 59.88~61.55%, 28.09~33.78%, and 2.99~5.28% methylation at total C, CG, CHG, and CHH sites, respectively. Shoot tips from dwarfing plants exhibited a hypermethylated pattern mostly due to increased CHH methylation, while leaves exhibited a hypomethylated pattern. According to GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis, DMGs (differentially methylated genes) and DEGs (differentially expressed genes) were enriched in hormone-related GO terms and KEGG pathways. Global correlation analysis between methylation and transcription revealed that mCpG in the gene body region enhanced gene expression and mCHH in the region near the TSS (transcription start site) was positively correlated with gene expression. Next, we found some hormone-related genes and TFs with significant changes in methylation and transcription, including SAURs, ARF, GA2ox, ABS1, bZIP, MYB, and NAC. This study presents a methylome map of the sweet cherry genome, revealed widespread DNA methylation alterations in scions caused by dwarfing rootstock, and obtained abundant genes with methylation and transcription alterations that are potentially involved in rootstock-induced growth changes in sweet cherry scions. Our findings can lay a good basis for further epigenetic studies on sweet cherry dwarfing and provide valuable new insight into understanding rootstock-scion interactions.
Keywords: DNA methylation; Prunus tomentosa; rootstock–scion interactions; transcriptome; whole-genome bisulfite sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
-
- World Flora Online (WFO) Prunus avium (L.) L. [(accessed on 28 July 2024)]. Available online: https://wfoplantlist.org/taxon/wfo-0001005630-2024-06?page=1.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

