Identification and Copy Number Variant Analysis of Enhancer Regions of Genes Causing Spinocerebellar Ataxia
- PMID: 39456985
- PMCID: PMC11508295
- DOI: 10.3390/ijms252011205
Identification and Copy Number Variant Analysis of Enhancer Regions of Genes Causing Spinocerebellar Ataxia
Abstract
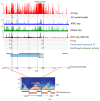
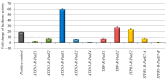
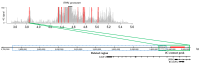
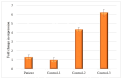
Currently, routine diagnostics for spinocerebellar ataxia (SCA) look for polyQ repeat expansions and conventional variations affecting the proteins encoded by known SCA genes. However, ~40% of the patients still remain without a genetic diagnosis after routine tests. Increasing evidence suggests that variations in the enhancer regions of genes involved in neurodegenerative disorders can also cause disease. Since the enhancers of SCA genes are not yet known, it remains to be determined whether variations in these regions are a cause of SCA. In this pilot project, we aimed to identify the enhancers of the SCA genes ATXN1, ATXN3, TBP and ITPR1 in the human cerebellum using 4C-seq, publicly available datasets, reciprocal 4C-seq, and luciferase assays. We then screened these enhancers for copy number variants (CNVs) in a cohort of genetically undiagnosed SCA patients. We identified two active enhancers for each of the four SCA genes. CNV analysis did not reveal any CNVs in the enhancers of the four SCA genes in the genetically undiagnosed SCA patients. However, in one patient, we noted a CNV deletion with an unknown clinical significance near one of the ITPR1 enhancers. These results not only reveal elements involved in SCA gene regulation but can also lead to the discovery of novel SCA-causing genetic variants. As enhancer variations are being increasingly recognized as a cause of brain disorders, screening the enhancers of ATXN1, ATXN3, TBP and ITPR1 for variations other than CNVs and identifying and screening enhancers of other SCA genes might elucidate the genetic cause in undiagnosed patients.
Keywords: 4C-seq; enhancer; genetic diagnosis; spinocerebellar ataxia.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Seixas A.I., Loureiro J.R., Costa C., Ordóñez-Ugalde A., Marcelino H., Oliveira C.L., Silveira I. A pentanucleotide ATTTC repeat insertion in the non-coding region of DAB1, mapping to SCA37, causes spinocerebellar ataxia. Am. J. Hum. Genet. 2017;101:87–103. doi: 10.1016/j.ajhg.2017.06.007. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

