Essential Genes Discovery in Microorganisms by Transposon-Directed Sequencing (Tn-Seq): Experimental Approaches, Major Goals, and Future Perspectives
- PMID: 39457080
- PMCID: PMC11508858
- DOI: 10.3390/ijms252011298
Essential Genes Discovery in Microorganisms by Transposon-Directed Sequencing (Tn-Seq): Experimental Approaches, Major Goals, and Future Perspectives
Abstract
Essential genes are crucial for microbial viability, playing key roles in both the primary and secondary metabolism. Since mutations in these genes can threaten organism viability, identifying them is challenging. Conditionally essential genes are required only under specific conditions and are important for functions such as virulence, immunity, stress survival, and antibiotic resistance. Transposon-directed sequencing (Tn-Seq) has emerged as a powerful method for identifying both essential and conditionally essential genes. In this review, we explored Tn-Seq workflows, focusing on eubacterial species and some yeast species. A comparison of 14 eubacteria species revealed 133 conserved essential genes, including those involved in cell division (e.g., ftsA, ftsZ), DNA replication (e.g., dnaA, dnaE), ribosomal function, cell wall synthesis (e.g., murB, murC), and amino acid synthesis (e.g., alaS, argS). Many other essential genes lack clear orthologues across different microorganisms, making them specific to each organism studied. Conditionally essential genes were identified in 18 bacterial species grown under various conditions, but their conservation was low, reflecting dependence on specific environments and microorganisms. Advances in Tn-Seq are expected to reveal more essential genes in the near future, deepening our understanding of microbial biology and enhancing our ability to manipulate microbial growth, as well as both the primary and secondary metabolism.
Keywords: Tn-Seq; essential genes; transposon.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


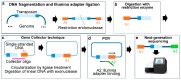
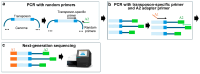
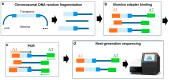

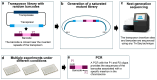

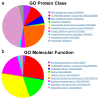

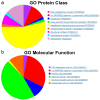
References
-
- Pugazhendhi A.S., Wei F., Hughes M., Coathup M. Musculoskeletal Infection. Springer; Cham, Switzerland: 2022. Bacterial adhesion, virulence, and biofilm formation; pp. 19–64.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

