Genome-Wide Identification of the Cyclic Nucleotide-Gated Ion Channel Gene Family and Expression Profiles Under Low-Temperature Stress in Luffa cylindrica L
- PMID: 39457112
- PMCID: PMC11508470
- DOI: 10.3390/ijms252011330
Genome-Wide Identification of the Cyclic Nucleotide-Gated Ion Channel Gene Family and Expression Profiles Under Low-Temperature Stress in Luffa cylindrica L
Abstract
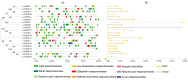
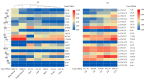
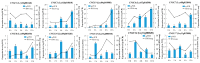
Cyclic nucleotide-gated ion channels (CNGCs) are cell membrane channel proteins for calcium ions. They have been reported to play important roles in survival and in the responses to environmental factors in various plants. However, little is known about the CNGC family and its functions in luffa (Luffa cylindrica L.). In this study, a bioinformatics-based method was used to identify members of the CNGC gene family in L. cylindrica. In total, 20 LcCNGCs were detected, and they were grouped into five subfamilies (I, II, Ⅲ, IV-a, and IV-b) in a phylogenetic analysis with CNGCs from Arabidopsis thaliana (20 AtCNGCs) and Momordica charantia (17 McCNGCs). The 20 LcCNGC genes were unevenly distributed on 11 of the 13 chromosomes in luffa, with none on Chromosomes 1 and 5. The members of each subfamily encoded proteins with highly conserved functional domains. An evolutionary analysis of CNGCs in luffa revealed three gene losses and a motif deletion. An examination of gene replication events during evolution indicated that two tandemly duplicated gene pairs were the primary driving force behind the evolution of the LcCNGC gene family. PlantCARE analyses of the LcCNGC promoter regions revealed various cis-regulatory elements, including those responsive to plant hormones (abscisic acid, methyl jasmonate, and salicylic acid) and abiotic stresses (light, drought, and low temperature). The presence of these cis-acting elements suggested that the encoded CNGC proteins may be involved in stress responses, as well as growth and development. Transcriptome sequencing (RNA-seq) analyses revealed tissue-specific expression patterns of LcCNGCs in various plant parts (roots, stems, leaves, flowers, and fruit) and the upregulation of some LcCNGCs under low-temperature stress. To confirm the accuracy of the RNA-seq data, 10 cold-responsive LcCNGC genes were selected for verification by quantitative real-time polymerase chain reaction (RT-qPCR) analysis. Under cold conditions, LcCNGC4 was highly upregulated (>50-fold increase in its transcript levels), and LcCNGC3, LcCNGC6, and LcCNGC13 were upregulated approximately 10-fold. Our findings provide new information about the evolution of the CNGC family in L. cylindrica and provide insights into the functions of the encoded CNGC proteins.
Keywords: CNGC; Luffa cylindrica L.; expression analysis; genome-wide identification; low temperature.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

