A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay
- PMID: 39457432
- PMCID: PMC11507897
- DOI: 10.3390/genes15101308
A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay
Abstract
Eukaryotic cells possess surveillance mechanisms that detect and degrade defective transcripts. Aberrant transcripts include mRNAs with a premature termination codon (PTC), targeted by the nonsense-mediated decay (NMD) pathway, and mRNAs lacking a termination codon, targeted by the nonstop decay (NSD) pathway. The eukaryotic exosome, a ribonucleolytic complex, plays a crucial role in mRNA processing and turnover through its catalytic subunits PM/Scl100 (Rrp6 in yeast), DIS3 (Rrp44 in yeast), and DIS3L1. Additionally, eukaryotic cells have other ribonucleases, such as SMG6 and XRN1, that participate in RNA surveillance. However, the specific pathways through which ribonucleases recognize and degrade mRNAs remain elusive. In this study, we characterized the involvement of human ribonucleases, both nuclear and cytoplasmic, in the mRNA surveillance mechanisms of NMD and NSD. We performed knockdowns of SMG6, PM/Scl100, XRN1, DIS3, and DIS3L1, analyzing the resulting changes in mRNA levels of selected natural NMD targets by RT-qPCR. Additionally, we examined the levels of different human β-globin variants under the same conditions: wild-type, NMD-resistant, NMD-sensitive, and NSD-sensitive. Our results demonstrate that all the studied ribonucleases are involved in the decay of certain endogenous NMD targets. Furthermore, we observed that the ribonucleases SMG6 and DIS3 contribute to the degradation of all β-globin variants, with an exception for βNS in the former case. This is also the case for PM/Scl100, which affects all β-globin variants except the NMD-sensitive variants. In contrast, DIS3L1 and XRN1 show specificity for β-globin WT and NMD-resistant variants. These findings suggest that eukaryotic ribonucleases are target-specific rather than pathway-specific. In addition, our data suggest that ribonucleases play broader roles in mRNA surveillance and degradation mechanisms beyond just NMD and NSD.
Keywords: mRNA degradation; mRNA surveillance; natural NMD targets; nonsense-mediated mRNA decay (NMD); nonstop decay (NSD); quality control.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
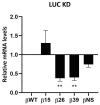
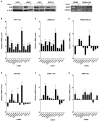
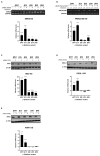
References
Publication types
MeSH terms
Substances
Grants and funding
- UIDB/04046/2020 (DOI: 10.54499/UIDB/04046/2020)/Fundação para a Ciência e a Tecnologia
- UIDP/04046/2020 (DOI: 10.54499/UIDP/04046/2020)/Fundação para a Ciência e a Tecnologia
- SFRH/BD/52495/2014/Fundação para a Ciência e a Tecnologia
- SFRH/BPD/98360/2013/Fundação para a Ciência e a Tecnologia
- UIDB/04612/2020/Fundação para a Ciência e a Tecnologia
LinkOut - more resources
Full Text Sources

