Use of a Novel Whole Blood Separation and Transport Device for Targeted and Untargeted Proteomics
- PMID: 39457630
- PMCID: PMC11504527
- DOI: 10.3390/biomedicines12102318
Use of a Novel Whole Blood Separation and Transport Device for Targeted and Untargeted Proteomics
Abstract
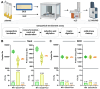
Background: There is significant interest in developing alternatives to traditional blood transportation and separation methods, which often require centrifugation and cold storage to preserve specimen integrity. Here we provide new performance findings that characterize a novel device that separates whole blood via lateral flow then dries the isolated components for room temperature storage and transport.
Methods: Untargeted proteomics was performed on non-small cell lung cancer (NSCLC) and normal healthy plasma applied to the device or prepared neat.
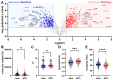
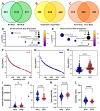
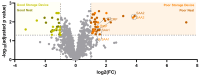
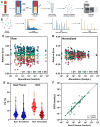
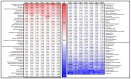
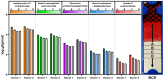
Results: Significantly, proteomic profiles from the storage device were more reproducible than from neat plasma. Proteins depleted or absent in the device preparation were shown to be absorbed onto the device membrane through largely hydrophilic interactions. Use of the device did not impact proteins relevant to an NSCLC clinical immune classifier. The device was also evaluated for use in targeted proteomics experiments using multiple-reaction monitoring (MRM) mass spectrometry. Intra-specimen detection intensity for protein targets between neat and device preparations showed a strong correlation, and device variation was comparable to the neat after normalization. Inter-specimen measurements between the device and neat preparations were also highly concordant.
Conclusions: These studies demonstrate that the lateral flow device is a viable blood separation and transportation tool for untargeted and targeted proteomics applications.
Keywords: LC-MS/MS; lateral flow; mass spectrometry; plasma proteomics.
Conflict of interest statement
Colin T. McDowell, Amanda L. Weaver, Nathan K. Keiser, Charles M. Nichols and Gary A. Pestano either are or were employed by Biodesix, Inc. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

