Taurine Protects against Silica Nanoparticle-Induced Apoptosis and Inflammatory Response via Inhibition of Oxidative Stress in Porcine Ovarian Granulosa Cells
- PMID: 39457890
- PMCID: PMC11506286
- DOI: 10.3390/ani14202959
Taurine Protects against Silica Nanoparticle-Induced Apoptosis and Inflammatory Response via Inhibition of Oxidative Stress in Porcine Ovarian Granulosa Cells
Abstract
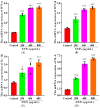
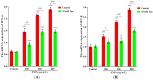
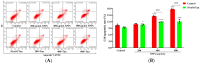
Silica nanoparticles (SNPs) induce reproductive toxicity through ROS production, which significantly limits their application. The protective effects of taurine (Tau) against SNP-induced reproductive toxicity remain unexplored. So this study aims to investigate the impact of Tau on SNP-induced porcine ovarian granulosa cell toxicity. In vitro, granulosa cells were exposed to SNPs combined with Tau. The localization of SNPs was determined by TEM. Cell viability was examined by CCK-8 assay. ROS levels were measured by CLSM and FCM. SOD and CAT levels were evaluated using ELISA and qPCR. Cell apoptosis was detected by FCM, and pro-inflammatory cytokine transcription levels were measured by qPCR. The results showed that SNPs significantly decreased cell viability, while increased cell apoptosis and ROS levels. Moreover, SOD and CAT were decreased, while IFN-α, IFN-β, IL-1β, and IL-6 were increased after SNP exposures. Tau significantly decreased intracellular ROS, while it increased SOD and CAT compared to SNPs alone. Additionally, Tau exhibited anti-inflammatory effects and inhibited cell apoptosis. On the whole, these findings suggest that Tau mitigates SNP-induced cytotoxicity by reducing oxidative stress, inflammatory response, and cell apoptosis. Tau may be an effective strategy to alleviate SNP-induced toxicity and holds promising application prospects in the animal husbandry and veterinary industry.
Keywords: apoptosis; granulosa cells; oxidative stress; silica nanoparticles; taurine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










Similar articles
-
Silica nanoparticles induce ovarian granulosa cell apoptosis via activation of the PERK-ATF4-CHOP-ERO1α pathway-mediated IP3R1-dependent calcium mobilization.Cell Biol Toxicol. 2023 Aug;39(4):1715-1734. doi: 10.1007/s10565-022-09776-4. Epub 2022 Nov 8. Cell Biol Toxicol. 2023. PMID: 36346508 Free PMC article.
-
Taurine alleviated acrylamide-induced ovarian toxicity via suppression of oxidative stress and apoptosis in mice.Toxicol Ind Health. 2025 May-Jun;41(5-6):282-293. doi: 10.1177/07482337251335541. Epub 2025 Apr 18. Toxicol Ind Health. 2025. PMID: 40250412
-
Resveratrol Attenuates Hydrogen Peroxide-induced Injury of Rat Ovarian Granulosa-lutein Cells by Resisting Oxidative Stress via the SIRT1/Nrf2/ARE Signaling Pathway.Curr Pharm Des. 2023;29(12):947-956. doi: 10.2174/1381612829666230403133322. Curr Pharm Des. 2023. PMID: 37013424
-
Protegrin-1 inhibits porcine ovarian granulosa cell apoptosis from H2O2-induced oxidative stress via the PERK/eIF2α/CHOP signaling pathway in vitro.Theriogenology. 2022 Feb;179:117-127. doi: 10.1016/j.theriogenology.2021.11.022. Epub 2021 Nov 29. Theriogenology. 2022. PMID: 34864562
-
Taurine effects on Bisphenol A-induced oxidative stress in the mouse testicular mitochondria and sperm motility.JBRA Assist Reprod. 2020 Oct 6;24(4):428-435. doi: 10.5935/1518-0557.20200017. JBRA Assist Reprod. 2020. PMID: 32550655 Free PMC article. Review.
References
-
- Jiang H.H., Zhang L., Wang X.J., Gu J., Song Z.L., Wei S.M., Guo H.H., Xu L., Qian X. Reductions in abundances of intracellular and extracellular antibiotic resistance genes by SiO2 nanoparticles during composting driven by mobile genetic elements. J. Environ. Manag. 2023;341:118071. doi: 10.1016/j.jenvman.2023.118071. - DOI - PubMed
-
- Li Y., Chen X., Jin R.H., Chen L., Dang M., Cao H., Dong Y., Cai B.L., Bai G., Gooding J.J., et al. Injectable hydrogel with MSNs/microRNA-21-5p delivery enables both immunomodification and enhanced angiogenesis for myocardial infarction therapy in pigs. Sci. Adv. 2021;7:eabd6740. doi: 10.1126/sciadv.abd6740. - DOI - PMC - PubMed
-
- Ferreira G.C., Sanches T.V.C., Mechler-Dreibi M.L., Almeida H.M.S., Storino G.Y., Sonalio K., Petri F.A.M., Martins T.S., da Silva L.C.C., Montassier H.J., et al. Efficacy evaluation of a novel oral silica-based vaccine in inducing mucosal immunity against Mycoplasma hyopneumoniae. Res. Vet. Sci. 2023;158:141–150. doi: 10.1016/j.rvsc.2023.03.018. - DOI - PubMed
-
- He J., Zhu T.Y., Mao N.N., Cai G.F., Gu P.F., Song Z.C., Lu X.Q., Yang Y., Wang D.Y. Cistanche deserticola polysaccharide-functionalized dendritic fibrous nano-silica as oral vaccine adjuvant delivery enhancing both the mucosal and systemic immunity. Int. J. Biol. Macromol. 2024;262:129982. doi: 10.1016/j.ijbiomac.2024.129982. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

