Novel Porcine Getah Virus from Diarrheal Piglets in Jiangxi Province, China: Prevalence, Genome Sequence, and Pathogenicity
- PMID: 39457910
- PMCID: PMC11503733
- DOI: 10.3390/ani14202980
Novel Porcine Getah Virus from Diarrheal Piglets in Jiangxi Province, China: Prevalence, Genome Sequence, and Pathogenicity
Abstract
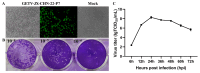
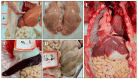
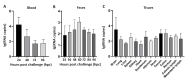
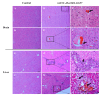
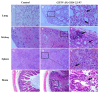
Getah virus (GETV) is a mosquito-borne virus belonging to the genus Alphavirus in the family Togaviridae. Its infection poses an increasing threat to animals and public health in China. In this study, an epidemiological survey of GETV on 46 pig farms in Jiangxi Province, China, was performed; GETV isolation and characterization were carried out, including a complete sequence determination and phylogenetic analysis; and pathogenicity of the GETV was experimentally investigated by inoculating newborn piglets with the isolated GETV strain. Epidemiological studies conducted on the organs of infected pigs, aborted piglets, and the blood of aborted sows sampled from pig farms in Jiangxi Province, China, demonstrated that 44 out of the 46 pig farms were positive for GETV, which is a positivity rate of 95.65% (44/46). Of the 411 samples tested, 47.93% (197/411) were found positive for GETV. A GETV strain called GETV-JX-CHN-22 was obtained, which showed stable proliferation in Vero cells. One-step growth curve results showed that the GETV-JX-CHN-22-P7 (passage 7) isolate reached a peak titer of 108.3 TCID50/mL at 24 hpi. An analysis of the whole-genome sequencing results showed that GETV-JX-CHN-22 (prototype) and GETV-JX-CHN-22-P7 shared nucleotide sequence similarities of 95.3% to 99.6% with 73 reference strains of GETV in GenBank. Genetic evolution analysis revealed that GETV-JX-CHN-22 and GETV-JX-CHN-22-P7 belonged to the GIII group, the same group members of most strains were reported in China. Animal inoculation experiments indicated that piglets exhibited typical symptoms and pathological changes of GETV infection after 24 h inoculation, which reproduced the pathogenicity of GETV field strain infections in piglets. To our knowledge, this study is the first report on the detection and isolation of porcine GETV associated with diarrhea from pig farms in Jiangxi Province, China. It is of great importance to study the infection spectrum, transmission mechanism, and public health significance of GETV. The results provide foundations for the genomic and biological (pathogenic) characteristics of the circulating GETV in Jiangxi Province, China.
Keywords: Getah virus; epidemiology; genomic sequencing; pathogenicity; swine; virus isolation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Li X.D., Qiu F.X., Yang H., Rao Y.N., Calisher C.H. Isolation of Getah virus from mosquitos collected on Hainan Island, China, and results of a serosurvey. Southeast Asian J. Trop. Med. Public Health. 1992;23:730–734. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

