A Longitudinal Study of Escherichia coli Clinical Isolates from the Tracheal Aspirates of a Paediatric Patient-Strain Type Similar to Pandemic ST131
- PMID: 39458299
- PMCID: PMC11509341
- DOI: 10.3390/microorganisms12101990
A Longitudinal Study of Escherichia coli Clinical Isolates from the Tracheal Aspirates of a Paediatric Patient-Strain Type Similar to Pandemic ST131
Abstract
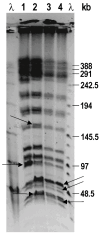
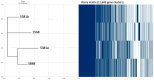
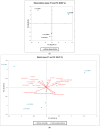
Escherichia coli is a Gram-negative bacterium and part of the intestinal microbiota. However, it can cause various diarrhoeal illnesses, i.e., traveller's diarrhoea, dysentery, and extraintestinal infections when the bacteria are translocated from the intestine to other organs, such as urinary tract infections, abdominal and pelvic infections, pneumonia, bacteraemia, and meningitis. It is also an important pathogen in intensive care units where cross-infection may cause intrahospital spread with serious consequences. Within this study, four E. coli isolates from the tracheal aspirates of a tracheotomised paediatric patient on chronic respiratory support were analysed and compared for antibiotic resistance and virulence potential. Genomes of all four isolates (5381a, 5381b, 5681, 5848) were sequenced using Oxford Nanopore Technology. According to PFGE analysis, the clones of isolates 5681 and 5848 were highly similar, and differ from 5381a and 5381b which were isolated first chronologically. All four E. coli isolates belonged to an unknown sequence type, related to the E. coli ST131, a pandemic clone that is evolving rapidly with increasing levels of antimicrobial resistance. All four E. coli isolates in this study exhibited a multidrug-resistant phenotype as, according to MIC data, they were resistant to ceftriaxone, ciprofloxacin, doxycycline, minocycline, and tetracycline. In addition, principal component analyses revealed that isolates 5681 and 5848, which were recovered later than 5381a and 5381b (two weeks and three weeks, respectively) possessed more complex antibiotic resistance genes and virulence profiles, which is concerning considering the short time period during which the strains were isolated.
Keywords: Escherichia coli; antimicrobial drug resistance; respiratory tract infections; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

