The Association between Resistance and Virulence of Klebsiella pneumoniae in High-Risk Clonal Lineages ST86 and ST101
- PMID: 39458306
- PMCID: PMC11509769
- DOI: 10.3390/microorganisms12101997
The Association between Resistance and Virulence of Klebsiella pneumoniae in High-Risk Clonal Lineages ST86 and ST101
Abstract
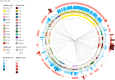
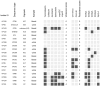
Klebsiella pneumoniae is an opportunistic pathogen known for two main pathotypes: classical K. pneumoniae (cKp), often multidrug-resistant and common in hospitals, and hypervirulent K. pneumoniae (hvKp), associated with severe community-acquired infections. The recent emergence of strains combining hypervirulence and resistance is alarming. This study investigates the distribution of sequence types (STs), resistance, and virulence factors in K. pneumoniae strains causing bloodstream and urinary tract infections in Croatia. In 2022, 200 consecutive K. pneumoniae isolates were collected from blood and urine samples across several Croatian hospitals. Whole genome sequencing was performed on 194 isolates. Within the analyzed K. pneumoniae population, the distribution of sequence types was determined with multi-locus sequence typing (MLST) and capsule loci, resistance, and virulence determinants were assessed with the bioinformatics tool Kleborate. The analysis identified 77 different STs, with ST101 (24.6%) being the most prevalent, predominantly linked to the K17 capsular type (CT), invasive device usage, high antimicrobial resistance, and low virulence scores. The highest virulence scores were recorded in ST86 isolates, which were predominantly linked to the K2 CT and included some strains with medium resistance scores. String tests were positive in 19 strains, but only four of those harbored hypermucoviscous genetic determinants. The most prevalent ST101 clone in Croatia demonstrated a diverging association between resistance and virulence. An alarming co-existence of resistance and virulence was recorded in the ST86 strains.
Keywords: Klebsiella pneumoniae; hypervirulence; resistance factors; virulence factors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Presence of hypervirulence-associated determinants in Klebsiella pneumoniae from hospitalised patients in Germany.Int J Med Microbiol. 2024 Mar;314:151601. doi: 10.1016/j.ijmm.2024.151601. Epub 2024 Feb 8. Int J Med Microbiol. 2024. PMID: 38359735
-
Comparative Analysis of Clinical and Genomic Characteristics of Hypervirulent Klebsiella pneumoniae from Hospital and Community Settings: Experience from a Tertiary Healthcare Center in India.Microbiol Spectr. 2022 Oct 26;10(5):e0037622. doi: 10.1128/spectrum.00376-22. Epub 2022 Aug 31. Microbiol Spectr. 2022. PMID: 36043878 Free PMC article.
-
[Investigation of Virulence Genes and Carbapenem Resistance Genes in Hypervirulent and Classical Isolates of Klebsiella pneumoniae Isolated from Various Clinical Specimens].Mikrobiyol Bul. 2023 Apr;57(2):188-206. doi: 10.5578/mb.20239915. Mikrobiyol Bul. 2023. PMID: 37067205 Turkish.
-
Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives.J Intern Med. 2020 Mar;287(3):283-300. doi: 10.1111/joim.13007. Epub 2019 Nov 21. J Intern Med. 2020. PMID: 31677303 Free PMC article. Review.
-
Global spread and evolutionary convergence of multidrug-resistant and hypervirulent Klebsiella pneumoniae high-risk clones.Pathog Glob Health. 2023 Jun;117(4):328-341. doi: 10.1080/20477724.2022.2121362. Epub 2022 Sep 11. Pathog Glob Health. 2023. PMID: 36089853 Free PMC article. Review.
Cited by
-
Whole-Genome Sequencing of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Isolated from Human Bloodstream Infections.Pathogens. 2025 Feb 20;14(3):205. doi: 10.3390/pathogens14030205. Pathogens. 2025. PMID: 40137690 Free PMC article.
-
Global molecular epidemiology of the incomplete CirA protein related to cefiderocol resistance in Klebsiella pneumoniae: a genome-based study.Microbiol Spectr. 2025 May 6;13(5):e0141024. doi: 10.1128/spectrum.01410-24. Epub 2025 Mar 19. Microbiol Spectr. 2025. PMID: 40105357 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

