Real-Time Monitoring of SARS-CoV-2 Variants in Oklahoma Wastewater through Allele-Specific RT-qPCR
- PMID: 39458310
- PMCID: PMC11509313
- DOI: 10.3390/microorganisms12102001
Real-Time Monitoring of SARS-CoV-2 Variants in Oklahoma Wastewater through Allele-Specific RT-qPCR
Abstract
During the COVID-19 pandemic, wastewater surveillance was used to monitor community transmission of SARS-CoV-2. As new genetic variants emerged, the need for timely identification of these variants in wastewater became an important focus. In response to increased reports of Omicron transmission across the United States, the Oklahoma Wastewater Surveillance team utilized allele-specific RT-qPCR assays to detect and differentiate variants, such as Omicron, from other variants found in wastewater in Oklahoma. The PCR assays showed presence of the Omicron variant in Oklahoma on average two weeks before official reports, which was confirmed through genomic sequencing of selected wastewater samples. Through continued surveillance from November 2021 to January 2022, we also demonstrated the transition from prevalence of the Delta variant to prevalence of the Omicron variant in local communities. We further assessed how this transition correlated with certain demographic factors characterizing each community. Our results highlight RT-qPCR assays as a rapid, simple, and cost-effective method for monitoring the community spread of SARS-CoV-2 genetic variants in wastewater. Additionally, they demonstrate that specific demographic factors such as ethnic composition and household income can correlate with the timing of SARS-CoV-2 variant introduction and spread.
Keywords: Omicron; RT-qPCR; SARS-CoV-2; early warning; sequencing; variant; wastewater surveillance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

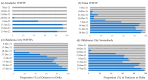
 and Delta
and Delta  variants in Oklahoma wastewater treatment plants (WWTP) and sewersheds, Oklahoma 2021–2022.
variants in Oklahoma wastewater treatment plants (WWTP) and sewersheds, Oklahoma 2021–2022.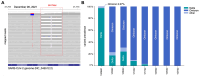


References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
-
- Tracking SARS-CoV-2 Variants. [(accessed on 5 January 2022)]. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants.
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

