Integrating the Transcriptome and Proteome to Postulate That TpiA and Pyk Are Key Enzymes Regulating the Growth of Mycoplasma Bovis
- PMID: 39458321
- PMCID: PMC11509987
- DOI: 10.3390/microorganisms12102012
Integrating the Transcriptome and Proteome to Postulate That TpiA and Pyk Are Key Enzymes Regulating the Growth of Mycoplasma Bovis
Abstract
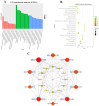
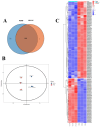
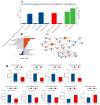
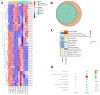
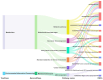
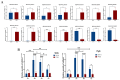
Mycoplasma bovis is a global problem for the cattle industry due to its high infection rates and associated morbidity, although its pathophysiology is poorly understood. In this study, the M. bovis transcriptome and proteome were analyzed to further investigate the biology of clinical isolates of M. bovis. A differential analysis of M. bovis, a clinical isolate (NX114), and an international type strain (PG45) at the logarithmic stage of growth, was carried out using prokaryotic transcriptome and 4D-label-free quantitative non-labeled proteomics. Transcriptomics and proteomics identified 193 DEGs and 158 DEPs, respectively, with significant differences in 49 proteins/34 transcriptomic CDS post-translational protein sequences (15 jointly up-regulated and 21 jointly down-regulated). GO comments indicate membrane, cytoplasmic and ribosome proteins were important components of the total proteins of M. bovis NX114 clinical isolate. KEGG enrichment revealed that M. bovis NX114 is mainly associated with energy metabolism, the biosynthesis of secondary metabolites, and the ABC transporters system. In addition, we annotated a novel adhesion protein that may be closely related to M. bovis infection. Triosephosphate isomerase (TpiA) and Pyruvate kinase (Pyk) genes may be the key enzymes that regulate the growth and maintenance of M. bovis and are involved in the pathogenic process as virulence factors. The results of the study revealed the biology of different isolates of M. bovis and may provide research ideas for the pathogenic mechanism of M. bovis.
Keywords: 4D-label-free quantitative proteomics; Mycoplasma bovis; Pyk; TpiA; prokaryotic transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
UHPLC/MS-Based Untargeted Metabolomics Reveals Metabolic Characteristics of Clinical Strain of Mycoplasma bovis.Microorganisms. 2023 Oct 21;11(10):2602. doi: 10.3390/microorganisms11102602. Microorganisms. 2023. PMID: 37894260 Free PMC article.
-
Ultra-High-Performance Liquid Chromatography-Orbitrap-MS-Based Untargeted Lipidomics Reveal Lipid Characteristics of a Clinical Strain of Mycoplasma bovis from Holstein Cow.Vet Sci. 2024 Nov 18;11(11):577. doi: 10.3390/vetsci11110577. Vet Sci. 2024. PMID: 39591351 Free PMC article.
-
MbovP0725, a secreted serine/threonine phosphatase, inhibits the host inflammatory response and affects metabolism in Mycoplasma bovis.mSystems. 2024 Apr 16;9(4):e0089123. doi: 10.1128/msystems.00891-23. Epub 2024 Mar 5. mSystems. 2024. PMID: 38440990 Free PMC article.
-
Proteomics analysis and its role in elucidation of functionally significant proteins in Mycoplasma bovis.Microb Pathog. 2017 Oct;111:50-59. doi: 10.1016/j.micpath.2017.08.024. Epub 2017 Aug 18. Microb Pathog. 2017. PMID: 28826762 Review.
-
Mycoplasma Bovis adhesins and their target proteins.Front Immunol. 2022 Oct 20;13:1016641. doi: 10.3389/fimmu.2022.1016641. eCollection 2022. Front Immunol. 2022. PMID: 36341375 Free PMC article. Review.
References
-
- Suwanruengsri M., Uemura R., Kanda T., Fuke N., Nueangphuet P., Pornthummawat A., Yasuda M., Hirai T., Yamaguchi R. Production of granulomas in Mycoplasma bovis infection associated with meningitis-meningoencephalitis, endocarditis, and pneumonia in cattle. J. Vet. Diagn. Investig. 2022;34:68–76. doi: 10.1177/10406387211053254. - DOI - PMC - PubMed
-
- Becker C.A.M., Ambroset C., Huleux A., Vialatte A., Colin A., Tricot A., Arcangioli M.A., Tardy F. Monitoring Mycoplasma bovis Diversity and Antimicrobial Susceptibility in Calf Feedlots Undergoing a Respiratory Disease Outbreak. Pathogens. 2020;9:593. doi: 10.3390/pathogens9070593. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

