The Identification of New Pharmacological Targets for the Treatment of Glaucoma: A Network Pharmacology Approach
- PMID: 39458974
- PMCID: PMC11509888
- DOI: 10.3390/ph17101333
The Identification of New Pharmacological Targets for the Treatment of Glaucoma: A Network Pharmacology Approach
Abstract
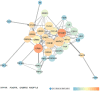
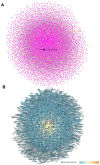
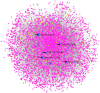
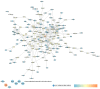
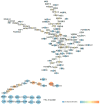
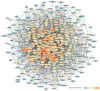
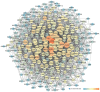
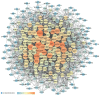
Background: Glaucoma is a progressive optic neuropathy characterized by the neurodegeneration and death of retinal ganglion cells (RGCs), leading to blindness. Current glaucoma interventions reduce intraocular pressure but do not address retinal neurodegeneration. In this effort, to identify new pharmacological targets for glaucoma management, we employed a network pharmacology approach. Methods: We first retrieved transcriptomic data from GEO, an NCBI database, and carried out GEO2R (an interactive web tool aimed at comparing two or more groups of samples in a GEO dataset). The GEO2R statistical analysis aimed at identifying the top differentially expressed genes (DEGs) and used these as input of STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) app within Cytoscape software, which builds networks of proteins starting from input DEGs. Analyses of centrality metrics using Cytoscape were carried out to identify nodes (genes or proteins) involved in network stability. We also employed the web-server software MIRNET 2.0 to build miRNA-target interaction networks for a re-analysis of the GSE105269 dataset, which reports analyses of microRNA expressions. Results: The pharmacological targets, identified in silico through analyses of the centrality metrics carried out with Cytoscape, were rescored based on correlations with entries in the PubMed and clinicaltrials.gov databases. When there was no match (82 out of 135 identified central nodes, in 8 analyzed networks), targets were considered "potential innovative" targets for the treatment of glaucoma, after further validation studies. Conclusions: Several druggable targets, such as GPCRs (e.g., 5-hydroxytryptamine 5A (5-HT5A) and adenosine A2B receptors) and enzymes (e.g., lactate dehydrogenase A or monoamine oxidase B), were found to be rescored as "potential innovative" pharmacological targets for glaucoma treatment.
Keywords: computational systems biology; glaucoma; network pharmacology; neuroinflammation; pharmacological targets.
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- GBD 2019 Blindness and Vision Impairment Collaborators. Vision Loss Expert Group of the Global Burden of Disease Study Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: The Right to Sight: An analysis for the Global Burden of Disease Study. Lancet Glob. Health. 2021;9:e144–e160. doi: 10.1016/S2214-109X(20)30489-7. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

