The Assembly of HTLV-1-How Does It Differ from HIV-1?
- PMID: 39459862
- PMCID: PMC11512237
- DOI: 10.3390/v16101528
The Assembly of HTLV-1-How Does It Differ from HIV-1?
Abstract
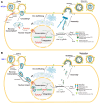
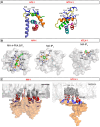
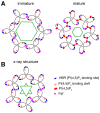
Retroviral assembly is a highly coordinated step in the replication cycle. The process is initiated when the newly synthesized Gag and Gag-Pol polyproteins are directed to the inner leaflet of the plasma membrane (PM), where they facilitate the budding and release of immature viral particles. Extensive research over the years has provided crucial insights into the molecular determinants of this assembly step. It is established that Gag targeting and binding to the PM is mediated by interactions of the matrix (MA) domain and acidic phospholipids such as phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2). This binding event, along with binding to viral RNA, initiates oligomerization of Gag on the PM, a process mediated by the capsid (CA) domain. Much of the previous studies have focused on human immunodeficiency virus type 1 (HIV-1). Although the general steps of retroviral replication are consistent across different retroviruses, comparative studies revealed notable differences in the structure and function of viral components. In this review, we present recent findings on the assembly mechanisms of Human T-cell leukemia virus type 1 and highlight key differences from HIV-1, focusing particularly on the molecular determinants of Gag-PM interactions and CA assembly.
Keywords: Gag polyprotein; capsid (CA); human T-cell leukemia virus type 1 (HTLV-1); human immunodeficiency virus type 1 (HIV-1); matrix (MA); phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2); plasma membrane (PM).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Smith R.G., Abrell J.W., Lewis B.J., Gallo R.C. Serological analysis of human deoxyribonucleic acid polymerases. Preparation and properties of antiserum to deoxyribonucleic acid polymerase I from human lymphoid cells. J. Biol. Chem. 1975;250:1702–1709. doi: 10.1016/S0021-9258(19)41750-X. - DOI - PubMed
-
- Poiesz B.J., Ruscetti F.W., Gazdar A.F., Bunn P.A., Minna J.D., Gallo R.C. Detection and isolation of type-C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA. 1980;75:7415–7419. doi: 10.1073/pnas.77.12.7415. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

