Comparison of Chikungunya Virus-Induced Disease Progression and Pathogenesis in Type-I Interferon Receptor-Deficient Mice (A129) and Two Wild-Type (129Sv/Ev and C57BL/6) Mouse Strains
- PMID: 39459867
- PMCID: PMC11512278
- DOI: 10.3390/v16101534
Comparison of Chikungunya Virus-Induced Disease Progression and Pathogenesis in Type-I Interferon Receptor-Deficient Mice (A129) and Two Wild-Type (129Sv/Ev and C57BL/6) Mouse Strains
Abstract
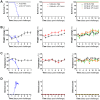
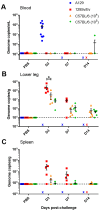
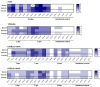
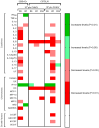
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus causing a debilitating febrile illness with rheumatic disease symptoms of arthralgia and arthritis. Since its spread outside of Africa in 2005, it continues to cause outbreaks and disseminates into new territories. Intervention strategies are urgently required, including vaccination and antiviral approaches. To test efficacy, the use of small animal models is required. Two mouse strains, A129, with a deficiency in their type-I interferon (IFN) receptor, and C57BL/6 are widely used. A direct comparison of these strains alongside the wild-type parental strain of the A129 mice, 129Sv/Ev, was undertaken to assess clinical disease progression, viral loads in key tissues, histological changes and levels of sera biomarkers. Our results confirm the severe disease course in A129 mice which was not observed in the parental 129Sv/Ev strain. Of the two wild-type strains, viral loads were higher in 129Sv/Ev mice compared to C57BL/6 counterparts. Our results have established these models and parameters for the future testing of vaccines and antiviral approaches.
Keywords: Chikungunya; model; mouse; pathogenesis; preclinical.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

