Molecular Detection by Rolling Circle Amplification Combined with Deep Sequencing of Mixed Infection by Bovine Papillomaviruses 2 and 4 in Carcinoma In Situ of the Bovine Esophageal Mucosa
- PMID: 39459892
- PMCID: PMC11512380
- DOI: 10.3390/v16101558
Molecular Detection by Rolling Circle Amplification Combined with Deep Sequencing of Mixed Infection by Bovine Papillomaviruses 2 and 4 in Carcinoma In Situ of the Bovine Esophageal Mucosa
Abstract
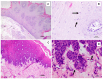
Papillomaviruses (PVs) are oncogenic and infect the skin and mucosa of various host species. Considering the recent advances in research on PVs using rolling circle amplification (RCA) followed by high-throughput sequencing (HTS), in this study, we aimed to investigate the bovine papillomavirus (BPV) types associated with proliferative lesions in the upper alimentary tract of an affected bull and characterize the viral strains through complete genome sequencing using this strategy. We analyzed the PV strains associated with two hyperplastic esophageal lesions through PCR using degenerate primer pairs and RCA, followed by HTS. HTS of the libraries generated using RCA products provided the whole genome sequence of BPV4 present in squamous papilloma, whereas the complete genome sequence of BPV2 and subgenomic fragments of BPV4 were identified in carcinoma in situ (CIS). For the first time, we have sequenced BPV2 identified from the CIS of the bovine upper alimentary canal. Additionally, RCA followed by HTS allowed characterization of the mixed infection by BPV2 and BPV4 in this lesion. These data reveal that BPV4 is not the only BPV type present in CIS of the esophageal mucous membrane; moreover, a mixed infection caused by BPV2 and BPV4 at the tested anatomical site was demonstrated.
Keywords: cattle; coinfection; esophagus; histopathology; next generation sequencing; papilloma; polymerase chain reaction; upper alimentary tract.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- ICTV International Committee on Taxonomy of Viruses. [(accessed on 15 March 2024)]. Available online: https://ictv.global.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

