The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains
- PMID: 39459895
- PMCID: PMC11512268
- DOI: 10.3390/v16101561
The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains
Abstract
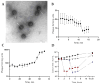
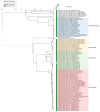
Bacteria of the Pseudomonas genus, including the Pseudomonas gessardii subgroup, play an important role in the environmental microbial communities. Psychrotolerant isolates of P. gessardii can produce thermostable proteases and lipases. When contaminating refrigerated raw milk, these bacteria spoil it by producing enzymes resistant to pasteurization. One possible way to prevent spoilage of raw milk is to use Pseudomonas lytic phages specific to undesirable P. gessardii isolates. The first phage, Pseudomonas vB_PseuGesM_254, was isolated and characterized, which is active against several proteolytic P. gessardii strains. This lytic myophage can infect and lyse its host strain at 24 °C and at low temperature (8 °C); so, it has the potential to prevent contamination of raw milk. The vB_PseuGesM_254 genome, 95,072 bp, shows a low level of intergenomic similarity with the genomes of known phages. Comparative proteomic ViPTree analysis indicated that vB_PseuGesM_254 is associated with a large group of Pseudomonas phages that are members of the Skurskavirinae and Gorskivirinae subfamilies and the Nankokuvirus genus. The alignment constructed using ViPTree shows that the vB_PseuGesM_254 genome has a large inversion between ~53,100 and ~70,700 bp, which is possibly a distinctive feature of a new taxonomic unit within this large group of Pseudomonas phages.
Keywords: Pseudomonas bacteriophage; Pseudomonas gessardii; environmental P. fluorescens complex; genome inversion; myovirus; proteolytic activity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

