Activation-Induced Marker Assay to Identify and Isolate HCV-Specific T Cells for Single-Cell RNA-Seq Analysis
- PMID: 39459954
- PMCID: PMC11512294
- DOI: 10.3390/v16101623
Activation-Induced Marker Assay to Identify and Isolate HCV-Specific T Cells for Single-Cell RNA-Seq Analysis
Abstract
Identification and isolation of antigen-specific T cells for downstream transcriptomic analysis is key for various immunological studies. Traditional methods using major histocompatibility complex (MHC) multimers are limited by the number of predefined immunodominant epitopes and MHC matching of the study subjects. Activation-induced markers (AIM) enable highly sensitive detection of rare antigen-specific T cells irrespective of the availability of MHC multimers. Herein, we have developed an AIM assay for the detection, sorting and subsequent single-cell RNA sequencing (scRNA-seq) analysis of hepatitis C virus (HCV)-specific T cells. We examined different combinations of the activation markers CD69, CD40L, OX40, and 4-1BB at 6, 9, 18 and 24 h post stimulation with HCV peptide pools. AIM+ CD4 T cells exhibited upregulation of CD69 and CD40L as early as 6 h post-stimulation, while OX40 and 4-1BB expression was delayed until 18 h. AIM+ CD8 T cells were characterized by the coexpression of CD69 and 4-1BB at 18 h, while the expression of CD40L and OX40 remained low throughout the stimulation period. AIM+ CD4 and CD8 T cells were successfully sorted and processed for scRNA-seq analysis examining gene expression and T cell receptor (TCR) usage. scRNA-seq analysis from this one subject revealed that AIM+ CD4 T (CD69+ CD40L+) cells predominantly represented Tfh, Th1, and Th17 profiles, whereas AIM+ CD8 T (CD69+ 4-1BB+) cells primarily exhibited effector and effector memory profiles. TCR analysis identified 1023 and 160 unique clonotypes within AIM+ CD4 and CD8 T cells, respectively. In conclusion, this approach offers highly sensitive detection of HCV-specific T cells that can be applied for cohort studies, thus facilitating the identification of specific gene signatures associated with infection outcome and vaccination.
Keywords: AIM assay; antigen-specific T cells; hepatitis C virus; single-cell RNA sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



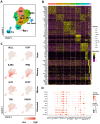
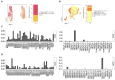
References
-
- Altosole T., Rotta G., Uras C.R.M., Bornheimer S.J., Fenoglio D. An optimized flow cytometry protocol for simultaneous detection of T cell activation induced markers and intracellular cytokines: Application to SARS-CoV-2 immune individuals. J. Immunol. Methods. 2023;515:113443. doi: 10.1016/j.jim.2023.113443. - DOI - PMC - PubMed
-
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

