Effects of Alkaline Solutions on the Structure and Function of Influenza A Virus
- PMID: 39459968
- PMCID: PMC11512367
- DOI: 10.3390/v16101636
Effects of Alkaline Solutions on the Structure and Function of Influenza A Virus
Abstract
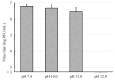
Influenza A virus (IAV) infection contributes to high annual morbidity and mortality, thus necessitating measures aimed at protecting against the disease. Alcohol-based disinfectants are commonly used to inactivate IAV, but they have several undesirable properties. In search of other means which would inactivate IAV, we focused on the effect of alkaline solutions on IAV. We found the viral infectivity remarkably decreased with treatment of an alkaline solution at pH 12.0 for 1 min, where destruction of the viral spikes was observed using an electron microscope. A more detailed examination revealed that the infectivity of IAV was remarkedly reduced by brief treatment with the alkaline solution at pH 11.75 or above, most likely due to the degradation of viral hemagglutinin protein. These results show that at a high pH, the haemagglutinin protein is degraded, resulting in very rapid inactivation of IAV.
Keywords: alkaline condition; hemagglutinin; hydrolysis; influenza A virus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Chemoreactive-Inspired Discovery of Influenza A Virus Dual Inhibitor to Block Hemagglutinin-Mediated Adsorption and Membrane Fusion.J Med Chem. 2020 Jul 9;63(13):6924-6940. doi: 10.1021/acs.jmedchem.0c00312. Epub 2020 Jun 26. J Med Chem. 2020. PMID: 32520560
-
Exploiting the Affimer platform against influenza A virus.mBio. 2024 Aug 14;15(8):e0180424. doi: 10.1128/mbio.01804-24. Epub 2024 Jul 22. mBio. 2024. PMID: 39037231 Free PMC article.
-
pH Optimum of Hemagglutinin-Mediated Membrane Fusion Determines Sensitivity of Influenza A Viruses to the Interferon-Induced Antiviral State and IFITMs.J Virol. 2017 May 12;91(11):e00246-17. doi: 10.1128/JVI.00246-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28356532 Free PMC article.
-
Influenza A virus entry inhibitors targeting the hemagglutinin.Viruses. 2013 Jan 22;5(1):352-73. doi: 10.3390/v5010352. Viruses. 2013. PMID: 23340380 Free PMC article. Review.
-
The role of fusion activity of influenza A viruses in their biological properties.Acta Virol. 2016 Jun;60(2):121-35. doi: 10.4149/av_2016_02_121. Acta Virol. 2016. PMID: 27265461 Review.
References
-
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

