The HIV-1 vpr R77Q Mutant Induces Apoptosis, G2 Cell Cycle Arrest, and Lower Production of Pro-Inflammatory Cytokines in Human CD4+ T Cells
- PMID: 39459974
- PMCID: PMC11512211
- DOI: 10.3390/v16101642
The HIV-1 vpr R77Q Mutant Induces Apoptosis, G2 Cell Cycle Arrest, and Lower Production of Pro-Inflammatory Cytokines in Human CD4+ T Cells
Abstract
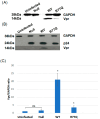
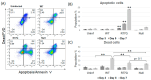
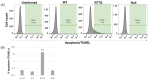
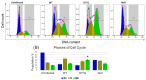
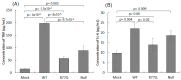
Acquired immunodeficiency syndrome (AIDS) occurs when HIV depletes CD4+ helper T cells. Some patients develop AIDS slowly or not at all, and are termed long-term non-progressors (LTNP), and while mutations in the HIV-1 Viral Protein R (vpr) gene such as R77Q are associated with LTNP, mechanisms for this correlation are unclear. This study examines the induction of apoptosis, cell cycle arrest, and pro-inflammatory cytokine release in the HUT78 T cell line following infection with replication-competent wild-type strain NL4-3, the R77Q mutant, or a vpr Null mutant. Our results show a significant enhancement of apoptosis and G2 cell cycle arrest in HUT78 cells infected with R77Q, but not with WT NL4-3 or the vpr Null strain. Conversely, HUT78 cells infected with the WT virus show higher levels of necrosis. We also detected lower TNF and IL-6 release after infection with R77Q vs. WT. The apoptotic phenotype was also seen in the CEM cell line and in primary CD4+ T cells. Protein expression of the R77Q vpr variant was low compared to WT vpr, but expression levels alone cannot explain these phenotypes because the Null virus did not show apoptosis or G2 arrest. These results suggest that R77Q triggers a non-inflammatory apoptotic pathway that attenuates inflammation, possibly contributing to LTNP.
Keywords: AIDS; HIV; apoptosis; immune activation; inflammation; necrosis; vpr.
Conflict of interest statement
We have no competing interests to declare.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

