The UCSC Genome Browser database: 2025 update
- PMID: 39460617
- PMCID: PMC11701590
- DOI: 10.1093/nar/gkae974
The UCSC Genome Browser database: 2025 update
Abstract
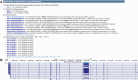
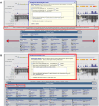
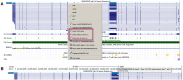
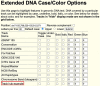
The UCSC Genome Browser (https://genome.ucsc.edu) is a widely utilized web-based tool for visualization and analysis of genomic data, encompassing over 4000 assemblies from diverse organisms. Since its release in 2001, it has become an essential resource for genomics and bioinformatics research. Annotation data available on Genome Browser includes both internally created and maintained tracks as well as custom tracks and track hubs provided by the research community. This last year's updates include over 25 new annotation tracks such as the gnomAD 4.1 track on the human GRCh38/hg38 assembly, the addition of three new public hubs, and significant expansions to the Genome Archive[GenArk) system for interacting with the enormous variety of assemblies. We have also made improvements to our interface, including updates to the browser graphic page, such as a new popup dialog feature that now displays item details without requiring navigation away from the main Genome Browser page. GenePred tracks have been upgraded with right-click options for zooming and precise navigation, along with enhanced mouseOver functions. Additional improvements include a new grouping feature for track hubs and hub description info links. A new tutorial focusing on Clinical Genetics has also been added to the UCSC Genome Browser.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Wagner N., Çelik M.H., Hölzlwimmer F.R., Mertes C., Prokisch H., Yépez V.A., Gagneur J.. Aberrant splicing prediction across human tissues. Nat. Genet. 2023; 55:861–870. - PubMed
MeSH terms
Grants and funding
- U01 CK000539/CK/NCEZID CDC HHS/United States
- DISC0-14514/California Institute for Regenerative Medicine
- OT2OD033761/NIH Office of the Director
- 6NU50CK000539-02-11/U.S. Department of Health and Human Services
- U24 HG002371/HG/NHGRI NIH HHS/United States
- 75D30121C11554/CC/CDC HHS/United States
- RF1 MH132662/MH/NIMH NIH HHS/United States
- RM1 HG011543/HG/NHGRI NIH HHS/United States
- R01MH120295/MH/NIMH NIH HHS/United States
- U24 MH132628/MH/NIMH NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- OT2 OD033761/OD/NIH HHS/United States
- U24HG002371/HG/NHGRI NIH HHS/United States
- 20-11088/California Department of Public Health

