Stacking correlation length in single-stranded DNA
- PMID: 39460618
- PMCID: PMC11602145
- DOI: 10.1093/nar/gkae934
Stacking correlation length in single-stranded DNA
Erratum in
-
Correction to 'Stacking correlation length in single-stranded DNA'.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf724. doi: 10.1093/nar/gkaf724. Nucleic Acids Res. 2025. PMID: 40682829 Free PMC article. No abstract available.
Abstract
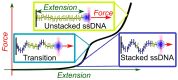
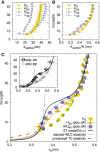
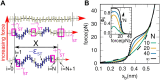
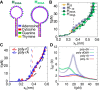
Base stacking is crucial in nucleic acid stabilization, from DNA duplex hybridization to single-stranded DNA (ssDNA) protein binding. While stacking energies are tiny in ssDNA, they are inextricably mixed with hydrogen bonding in DNA base pairing, making their measurement challenging. We conduct unzipping experiments with optical tweezers of short poly-purine (dA and alternating dG and dA) sequences of 20-40 bases. We introduce a helix-coil model of the stacking-unstacking transition that includes finite length effects and reproduces the force-extension curves. Fitting the model to the experimental data, we derive the stacking energy per base, finding the salt-independent value $\Delta G_0^{ST}=0.14(3)$ kcal/mol for poly-dA and $\Delta G_0^{ST}=0.07(3)$ kcal/mol for poly-dGdA. Stacking in these polymeric sequences is predominantly cooperative with a correlation length of ∼4 bases at zero force . The correlation length reaches a maximum of ∼10 and 5 bases at the stacking-unstacking transition force of ∼10 and 20 pN for poly-dA and poly-dGdA, respectively. The salt dependencies of the cooperativity parameter in ssDNA and the energy of DNA hybridization are in agreement, suggesting that double-helix stability is primarily due to stacking. Analysis of poly-rA and poly-rC RNA sequences shows a larger stacking stability but a lower stacking correlation length of ∼2 bases.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Saenger W. Principles of Nucleic Acid Structure: Chapters 1, 4 and 6. 2013; NY: Springer Verlag, Springer New York.
-
- Holbrook J.A., Capp M.W., Saecker R.M., Record M.T. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: Interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry. 1999; 38:8409–8422. - PubMed
-
- Kim C., Wold M.S. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995; 34:2058–2064. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

