A de novo missense mutation in PPP2R5D alters dopamine pathways and morphology of iPSC-derived midbrain neurons
- PMID: 39460716
- PMCID: PMC11811633
- DOI: 10.1093/stmcls/sxae068
A de novo missense mutation in PPP2R5D alters dopamine pathways and morphology of iPSC-derived midbrain neurons
Abstract
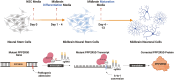
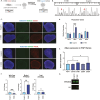
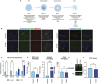
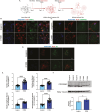
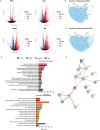
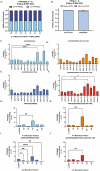
Induced pluripotent stem cell (iPSC) models of neurodevelopmental disorders (NDDs) have promoted an understanding of commonalities and differences within or across patient populations by revealing the underlying molecular and cellular mechanisms contributing to disease pathology. Here, we focus on developing a human model for PPP2R5D-related NDD, called Jordan syndrome, which has been linked to Early-Onset Parkinson's Disease (EOPD). Here we sought to understand the underlying molecular and cellular phenotypes across multiple cell states and neuronal subtypes in order to gain insight into Jordan syndrome pathology. Our work revealed that iPSC-derived midbrain neurons from Jordan syndrome patients display significant differences in dopamine-associated pathways and neuronal architecture. We then evaluated a CRISPR-based approach for editing heterozygous dominant G-to-A mutations at the transcript level in patient-derived neural stem cells. Our findings show that site-directed RNA editing is influenced by sgRNA length and cell type. These studies support the potential for a CRISPR RNA editor system to selectively edit mutant transcripts harboring G-to-A mutations in neural stem cells while providing an alternative editing technology for those suffering from NDDs.
Keywords: EOPD; Jordans Syndrome; NDD; RNA editing and CRISPR/Cas13; disease modeling; iPSC-derived neuron; midbrain neuron; stem cell.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Marrus N, Hall L.. Intellectual disability and language disorder. Child Adolesc Psychiatr Clin N Am. 2017;26:539-554. https://doi.org/10.1016/j.chc.2017.03.001 - DOI - PMC - PubMed
-
- Jamra R. Genetics of autosomal recessive intellectual disability. Medizinische Genetik : Mitteilungsblatt des Berufsverbandes Medizinische Genetik e.V. 2018;30:323-327. https://doi.org/10.1007/s11825-018-0209-z - DOI - PMC - PubMed
-
- Järvelä I, Määttä T, Acharya A, et al.Exome sequencing reveals predominantly de novo variants in disorders with intellectual disability (ID) in the founder population of Finland. Hum Genet. 2021;140:1011-1029. https://doi.org/10.1007/s00439-021-02268-1 - DOI - PMC - PubMed
-
- Chen S, Fragoza R, Klei L, et al.An interactome perturbation framework prioritizes damaging missense mutations for developmental disorders. Nat Genet. 2018;50:1032-1040. https://doi.org/10.1038/s41588-018-0130-z - DOI - PMC - PubMed
-
- Shang L, Henderson LB, Cho MT, et al.De novo missense variants in PPP2R5D are associated with intellectual disability, macrocephaly, hypotonia, and autism. Neurogenetics. 2016;17:43-49. https://doi.org/10.1007/s10048-015-0466-9 - DOI - PMC - PubMed

