Specialized contact sites regulate the fusion of chlamydial inclusion membranes
- PMID: 39461996
- PMCID: PMC11513123
- DOI: 10.1038/s41467-024-53443-7
Specialized contact sites regulate the fusion of chlamydial inclusion membranes
Erratum in
-
Author Correction: Specialized contact sites regulate the fusion of chlamydial inclusion membranes.Nat Commun. 2025 Feb 7;16(1):1454. doi: 10.1038/s41467-025-56777-y. Nat Commun. 2025. PMID: 39920215 Free PMC article. No abstract available.
Abstract
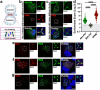
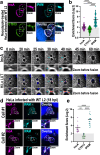
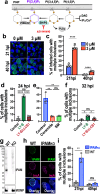
The intracellular bacterial pathogen Chlamydia trachomatis replicates within a membrane-bound compartment called the inclusion. Upon infection with several chlamydiae, each bacterium creates its own inclusion, resulting in multiple inclusions within each host cell. Ultimately, these inclusions fuse together in a process that requires the chlamydial protein IncA. Here, we show that inclusions form unique contact sites (inclusion contact sites, ICSs) prior to fusion, that serve as fusogenic platforms in which specific lipids and chlamydial proteins concentrate. Fusion depends on IncA clustering within ICSs and is regulated by PI(3,4)P2 and sphingolipids. As IncA concentrates within ICSs, its C-terminus likely interacts in trans with IncA on the apposing membrane, securing a high concentration of IncA at fusion sites. This regulatory mechanism contrasts with eukaryotic or viral fusion systems that are either composed of multiple proteins or use a change in pH to initiate membrane fusion. Thus, our study demonstrates that Chlamydia-mediated membrane fusion is primarily regulated by specific structural domains in IncA and its local organization on the inclusion membrane, which is affected by the host cell lipid composition.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

