CRISPR-Cas system positively regulates virulence of Salmonella enterica serovar Typhimurium
- PMID: 39462402
- PMCID: PMC11514906
- DOI: 10.1186/s13099-024-00653-5
CRISPR-Cas system positively regulates virulence of Salmonella enterica serovar Typhimurium
Abstract
Background: Salmonella, a foodborne pathogen, possesses a type I-E clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated (Cas) system. We investigated the system's role in regulating Salmonella virulence by deleting the CRISPR arrays and Cas operon.
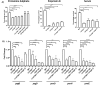
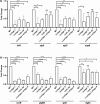
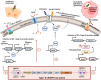
Results: Our study demonstrates invasion and proliferation defects of CRISPR-Cas knockout strains in intestinal epithelial cells and macrophages owing to the repression of invasion and virulence genes. However, proliferation defects were not observed in the Gp91phox-/- macrophages, suggesting the system's role in the pathogens' antioxidant defense. We deduced that the CRISPR-Cas system positively regulates H2O2 importer (OmpW), catalase (katG), peroxidase (ahpC), and superoxide dismutase (soda and sodCI), thereby protecting the cells from oxidative radicals. The knockout strains were attenuated in in-vivo infection models (Caenorhabditis elegans and BALB/c mice) due to hypersensitivity against antimicrobial peptides, complement proteins, and oxidative stress. The attenuation in virulence was attributed to the suppression of LPS modifying (pmr) genes, antioxidant genes, master regulators, and effectors of the SPI-1 (invasion) and SPI-2 (proliferation) islands in knockout strains. The regulation could be attributed to the partial complementarity of the CRISPR spacers with these genes.
Conclusions: Overall, our study extends our understanding of the role of the CRISPR-Cas system in Salmonella pathogenesis and its virulence determinants.
Keywords: Salmonella; Anti-oxidant genes; SPI-1-T3SS; SPI-2-T3SS; Type 1-E CRISPR-Cas system; Virulence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Sampson TR, Weiss DS. Cas9-dependent endogenous gene regulation is required for bacterial virulence. Biochem Soc Trans. 2013;41:1407–11. 10.1042/BST20130163. - PubMed
-
- Louwen R, Horst-Kreft D, De Boer AG, Van Der Graaf L, De Knegt G, Hamersma M, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur J Clin Microbiol Infect Dis. 2013;32:207–26. 10.1007/S10096-012-1733-4. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

