S-RBD-modified and miR-486-5p-engineered exosomes derived from mesenchymal stem cells suppress ferroptosis and alleviate radiation-induced lung injury and long-term pulmonary fibrosis
- PMID: 39462403
- PMCID: PMC11515248
- DOI: 10.1186/s12951-024-02830-9
S-RBD-modified and miR-486-5p-engineered exosomes derived from mesenchymal stem cells suppress ferroptosis and alleviate radiation-induced lung injury and long-term pulmonary fibrosis
Abstract
Background: Radiation-induced lung injury (RILI) is associated with alveolar epithelial cell death and secondary fibrosis in injured lung. Mesenchymal stem cell (MSC)-derived exosomes have regenerative effect against lung injury and the potential to intervene of RILI. However, their intervention efficacy is limited because they lack lung targeting characters and do not carry sufficient specific effectors. SARS-CoV-2 spike glycoprotein (SARS-CoV-2-S-RBD) binds angiotensin-converting enzyme 2 (ACE2) receptor and mediates interaction with host cells. MiR-486-5p is a multifunctional miRNA with angiogenic and antifibrotic potential and acts as an effector in MSC-derived exosomes. Ferroptosis is a form of cell death associated with radiation injury, its roles and mechanisms in RILI remain unclear. In this study, we developed an engineered MSC-derived exosomes with SARS-CoV-2-S-RBD- and miR-486-5p- modification and investigated their intervention effects on RIPF and action mechanisms via suppression of epithelial cell ferroptosis.
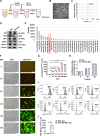
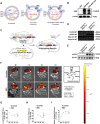
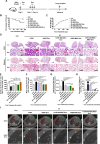
Results: Adenovirus-mediated gene modification led to miR-486-5p overexpression in human umbilical cord MSC exosomes (p < 0.05), thereby constructing miR-486-5p engineered MSC exosomes (miR-486-MSC-Exo). MiR-486-MSC-Exo promoted the proliferation and migration of irradiated mouse lung epithelial (MLE-12) cells in vitro and inhibited RILI in vivo (all p < 0.05). MiR-486-MSC-Exo suppressed ferroptosis in MLE-12 cells, and an in vitro assay revealed that the expression of fibrosis-related genes is up-regulated following ferroptosis (both p < 0.05). MiR-486-MSC-Exo reversed the up-regulated expression of fibrosis-related genes induced by TGF-β1 in vitro and improved pathological fibrosis in RIPF mice in vivo (all p < 0.05). SARS-CoV-2-S-RBD-modified and miR-486-5p-engineered MSC exosomes (miR-486-RBD-MSC-Exo) were also constructed, and the distribution of DiR dye-labeled miR-486-RBD-MSC-Exo in hACE2CKI/CKI Sftpc-Cre+ mice demonstrated long-term retention in the lung (p < 0.05). MiR-486-RBD-MSC-Exo significantly improved the survival rate and pathological changes in hACE2CKI/CKI Sftpc-Cre+ RIPF mice (all p < 0.05). Furthermore, miR-486-MSC-Exo exerted anti-fibrotic effects via targeted SMAD2 inhibition and Akt phosphorylation activation (p < 0.05).
Conclusions: Engineered MSC exosomes with SARS-CoV-2-S-RBD- and miR-486-5p-modification were developed. MiR-486-RBD-MSC-Exo suppressed ferroptosis and fibrosis of MLE-12 cells in vitro, and alleviated RILI and long-term RIPF in ACE2 humanized mice in vivo. MiR-486-MSC-Exo exerted anti-fibrotic effects via SMAD2 inhibition and Akt activation. This study provides a potential approach for RIPF intervention.
Keywords: Engineered exosomes; Ferroptosis; Mesenchymal stem cells; MiR-486-5p; Pulmonary fibrosis; Radiation-induced pulmonary injury; SARS-CoV-2-S-RBD.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

