Salivary Extracellular Vesicles Separation: Analysis of Ultracentrifugation-Based Protocols
- PMID: 39462790
- PMCID: PMC12021316
- DOI: 10.1111/odi.15171
Salivary Extracellular Vesicles Separation: Analysis of Ultracentrifugation-Based Protocols
Abstract
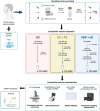
Introduction: The clinical potential of extracellular vesicles (EVs) is widely acknowledged, yet the standardization and reproducibility of its separation remain challenging. This study compares three protocols: ultracentrifugation (UC), UC with purification step (UC + PS), and a combined protocol using polymer-based precipitation and UC (PBP + UC).
Methods: Salivary samples were collected from healthy donors. EVs were separated (UC, UC + PS, and PBP + UC) and characterized using transmission electron microscopy, nanoparticle tracking analysis, EV purity, RNA concentration, and Western blotting. miRNA expression was evaluated by quantitative RT-PCR. Statistical analyses comparing groups were performed using ANOVA.
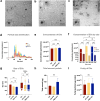
Results: All methods successfully separated CD9+ and CD63+ EVs from saliva. The UC + PS and PBP + UC protocols yielded the highest concentrations of EVs, enriched in < 200 nm vesicles. EV purity and RNA recovery were comparable among all methods. Expression of miR-16, miR-27a, and miR-99a was successfully detected using all methods.
Conclusions: The UC + PS and PBP + UC protocols demonstrate comparable efficiency in separating salivary EVs. However, the combined PBP + UC protocol, with its simplified processing capability, offers a significant advantage, particularly in the initial phase of EV separation. This finding suggests its potential application in clinical settings where time-sensitive simple processing is critical. Further validation is needed to confirm its effectiveness for transcriptomic and proteomic analyses.
Keywords: PEG; extracellular vesicles; miRNAs; saliva; ultracentrifugation.
© 2024 The Author(s). Oral Diseases published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Aqrawi, L. A. , Galtung H. K., Vestad B., et al. 2017. “Identification of Potential Saliva and Tear Biomarkers in Primary Sjögren's Syndrome, Utilising the Extraction of Extracellular Vesicles and Proteomics Analysis.” Arthritis Research and Therapy 19, no. 1: 1–15. 10.1186/s13075-017-1228-x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

