Cracking Lysine Crotonylation (Kcr): Enlightening a Promising Post-Translational Modification
- PMID: 39462860
- PMCID: PMC11776371
- DOI: 10.1002/cbic.202400639
Cracking Lysine Crotonylation (Kcr): Enlightening a Promising Post-Translational Modification
Abstract
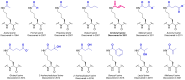
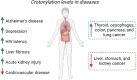
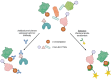
Lysine crotonylation (Kcr) is a recently discovered post-translational modification (PTM). Both histone and non-histone Kcr-proteins have been associated with numerous diseases including cancer, acute kidney injury, HIV latency, and cardiovascular disease. Histone Kcr enhances gene expression to a larger extend than the extensively studied lysine acetylation (Kac), suggesting Kcr as a novel potential therapeutic target. Although numerous scientific reports on crotonylation were published in the last years, relevant knowledge gaps concerning this PTM and its regulation still remain. To date, only few selective Kcr-interacting proteins have been identified and selective methods for the enrichment of Kcr-proteins in chemical proteomics analysis are still lacking. The development of new techniques to study this underexplored PTM could then clarify its function in health and disease and hopefully accelerate the development of new therapeutics for Kcr-related disease. Herein we briefly review what is known about the regulation mechanisms of Kcr and the current methods used to identify Kcr-proteins and their interacting partners. This report aims to highlight the significant potential of Kcr as a therapeutic target and to identify the existing scientific gaps that new research must address.
Keywords: Chemical proteomics; Crotonyl-CoA; Enzymatic PTMs regulation; Post-translational modifications (PTMs).
© 2024 The Author(s). ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

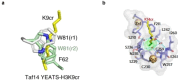




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

