CircMALAT1 promotes the proliferation and metastasis of intrahepatic cholangiocarcinoma via the miR-512-5p/VCAM1 axis
- PMID: 39463204
- PMCID: PMC11877140
- DOI: 10.3724/abbs.2024185
CircMALAT1 promotes the proliferation and metastasis of intrahepatic cholangiocarcinoma via the miR-512-5p/VCAM1 axis
Abstract
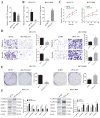
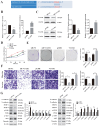
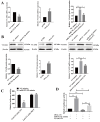
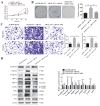
Circular RNAs play a pivotal role in the progression of various cancers. In our previous study, we observed high expression of the circRNA MALAT1 (cMALAT1) in intrahepatic cholangiocarcinoma (ICC) cells co-incubated with activated hepatic stellate cells. This study is designed to explore the roles of cMALAT1 and the underlying mechanisms in ICC. We find that cMALAT1 significantly facilitates the progression of ICC both in vitro and in vivo. The binding between cMALAT1 and miR-512-5p is subsequently confirmed through RNA pull-down experiments. As anticipated, the application of miR-512-5p mimics noticeably reverses the cMALAT1 overexpression-induced malignant phenotypes of ICC cells. Furthermore, VCAM1 is identified as a downstream gene of the cMALAT1/miR-512-5p axis. Importantly, silencing of VCAM1 not only effectively suppresses the malignant phenotypes of ICC cells but also significantly impairs the functions of cMALAT1. Our study reveals that cMALAT1 promotes the progression of ICC by competitively binding to VCAM1 mRNA with miR-512-5p, leading to the upregulation of VCAM1 expression and the activation of the PI3K/AKT signaling pathway.
Keywords: EMT; ICC; VCAM1; circular MALAT1; miR-512-5p.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. . 2022;72:7–33. doi: 10.3322/caac.21708. - DOI - PubMed
-
- Beal EW, Tumin D, Moris D, Zhang XF, Chakedis J, Dilhoff M, Schmidt CM, et al. Cohort contributions to trends in the incidence and mortality of intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr. . 2018;7:270–276. doi: 10.21037/hbsn.2018.03.16. - DOI - PMC - PubMed
-
- Sabbatino F, Villani V, Yearley JH, Deshpande V, Cai L, Konstantinidis IT, Moon C, et al. PD-L1 and HLA class I antigen expression and clinical course of the disease in intrahepatic cholangiocarcinoma. Clin Cancer Res. . 2016;22:470–478. doi: 10.1158/1078-0432.CCR-15-0715. - DOI - PMC - PubMed
-
- Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, Huang N, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. . 2019;18:131. doi: 10.1186/s12943-019-1056-5. - DOI - PMC - PubMed
-
- Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. . 2019;18:119. doi: 10.1186/s12943-019-1046-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

