Expression of lipid-metabolism genes is correlated with immune microenvironment and predicts prognosis of hepatocellular carcinoma
- PMID: 39463421
- PMCID: PMC11514195
- DOI: 10.1038/s41598-024-76578-5
Expression of lipid-metabolism genes is correlated with immune microenvironment and predicts prognosis of hepatocellular carcinoma
Abstract
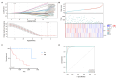
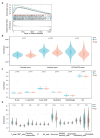
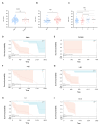
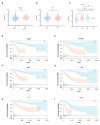
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors. This study was aimed to identify a lipid metabolism-related signature associated with the HCC microenvironment to improve the prognostic prediction of HCC patients. Clinical information and expression profile data were downloaded from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases, including the GEO dataset GSE76427. The gene expression profile of lipid metabolism was downloaded from Molecular Signatures Database (MSigDB) database. The infiltrating immune cells were estimated by the Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE), MCP-counter, and TIMER algorithms. Functional analysis, including Gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene set enrichment analysis (GSEA) were performed to elucidate the underlying mechanisms. The prognostic risk model was performed by Least absolute shrinkage and selection operator (LASSO) algorithm and Cox regression analysis. Two distinct subgroups of survival were identified. Better prognosis was associated with high immune score, high abundance of immune infiltrating cells, and high immune status. GO and KEGG analysis showed that differentially expressed genes (DEGs) between the two subgroups were mainly enriched in immune related pathways. GSEA analysis suggested that the expression of lipid metabolism related genes (LMRGs) was related to dysregulation of immune in the high-risk group. Risk models and clinical features based on LMRGs predicted HCC prognosis. This study indicated that the lipid metabolism-related signature was important for the prognosis of HCC. The expression of LMRGs was related to the immune microenvironment of HCC patients and could be used to predict the prognosis of HCC.
Keywords: Hepatocellular carcinoma; LMRGs; Lipid metabolism; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

