This is a preprint.
Chromatin interaction maps of human arterioles reveal new mechanisms for the genetic regulation of blood pressure
- PMID: 39463975
- PMCID: PMC11507733
- DOI: 10.1101/2024.10.09.617511
Chromatin interaction maps of human arterioles reveal new mechanisms for the genetic regulation of blood pressure
Update in
-
Chromatin interaction maps of human arterioles reveal mechanisms for the genetic regulation of blood pressure.Nat Commun. 2025 Jul 17;16(1):6577. doi: 10.1038/s41467-025-61656-7. Nat Commun. 2025. PMID: 40675959 Free PMC article.
Abstract
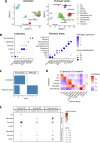
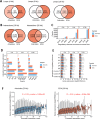
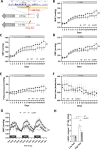
Arterioles are small blood vessels located just upstream of capillaries in nearly all tissues. The constriction and dilation of arterioles regulate tissue perfusion and are primary determinants of systemic blood pressure (BP). Abnormalities in arterioles are central to the development of major diseases such as hypertension, stroke, and microvascular complications of diabetes. Despite the broad and essential role of arterioles in physiology and disease, current knowledge of the functional genomics of arterioles is largely absent, partly because it is challenging to obtain and analyze human arteriole samples. Here, we report extensive maps of chromatin interactions, single-cell expression, and other molecular features in human arterioles and uncover new mechanisms linking human genetic variants to gene expression in vascular cells and the development of hypertension. Compared to large arteries, arterioles exhibited a higher proportion of pericytes which were strongly associated with BP traits. BP-associated single nucleotide polymorphisms (SNPs) were enriched in chromatin interaction regions in arterioles, particularly through enhancer SNP-promoter interactions, which were further linked to gene expression specificity across tissue components and cell types. Using genomic editing in animal models and human induced pluripotent stem cells, we discovered novel mechanisms linking BP-associated noncoding SNP rs1882961 to gene expression through long-range chromatin contacts and revealed remarkable effects of a 4-bp noncoding genomic segment on hypertension in vivo. We anticipate that our rich data and findings will advance the study of the numerous diseases involving arterioles. Moreover, our approach of integrating chromatin interaction mapping in trait-relevant tissues with SNP analysis and in vivo and in vitro genome editing can be applied broadly to bridge the critical gap between genetic discoveries and physiological understanding.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
