This is a preprint.
Methylation Clocks Do Not Predict Age or Alzheimer's Disease Risk Across Genetically Admixed Individuals
- PMID: 39464059
- PMCID: PMC11507840
- DOI: 10.1101/2024.10.16.618588
Methylation Clocks Do Not Predict Age or Alzheimer's Disease Risk Across Genetically Admixed Individuals
Abstract
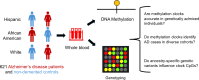
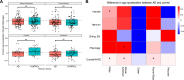
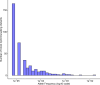
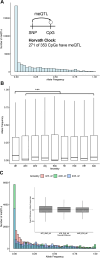
Epigenetic clocks that quantify rates of aging from DNA methylation patterns across the genome have emerged as a potential biomarker for risk of age-related diseases, like Alzheimer's disease (AD), and environmental and social stressors. However, methylation clocks have not been validated in genetically diverse cohorts. Here we evaluate a set of methylation clocks in 621 AD patients and matched controls from African American, Hispanic, and white cohorts. The clocks are less accurate at predicting age in genetically admixed individuals, especially those with substantial African ancestry, than in the white cohort. The clocks also do not consistently identify age acceleration in admixed AD cases compared to controls. Methylation QTL (meQTL) commonly influence CpGs in clocks, and these meQTL have significantly higher frequencies in African genetic ancestries. Our results demonstrate that methylation clocks often fail to predict age and AD risk beyond their training populations and suggest avenues for improving their portability.
Conflict of interest statement
4.11Competing interestsre The authors declare no competing interests.
Figures

References
-
- Banovich Nicholas E. et al. (Sept. 2014). “Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels”. en. In: PLoS Genetics 10.9. Ed. by Reddy Timothy E., e1004663. issn: 1553–7404. doi: 10.1371/journal.pgen.1004663. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
