This is a preprint.
A fitness distribution law for amino-acid replacements
- PMID: 39464166
- PMCID: PMC11507765
- DOI: 10.1101/2024.10.11.617952
A fitness distribution law for amino-acid replacements
Abstract
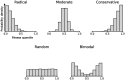
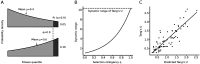
The effect of replacing the amino acid at a given site in a protein is difficult to predict. Yet, evolutionary comparisons have revealed highly regular patterns of interchangeability between pairs of amino acids, and such patterns have proved enormously useful in a range of applications in bioinformatics, evolutionary inference, and protein design. Here we reconcile these apparently contradictory observations using fitness data from over 350,000 experimental amino acid replacements. Almost one-quarter of the 20 × 19 = 380 types of replacements have broad distributions of fitness effects (DFEs) that closely resemble the background DFE for random changes, indicating an overwhelming influence of protein context in determining mutational effects. However, we also observe that the 380 pair-specific DFEs closely follow a maximum entropy distribution, specifically a truncated exponential distribution. The shape of this distribution is determined entirely by its mean, which is equivalent to the chance that a replacement of the given type is fitter than a random replacement. In this type of distribution, modest deviations in the mean correspond to much larger changes in the probability of falling in the far right tail, so that modest differences in mean exchangeability may result in much larger differences in the chance of a highly fit mutation. Indeed, we show that under the assumption that purifying selection filters out the vast majority of mutations, the maximum entropy distributions of fitness effects inferred from deep mutational scanning experiments predict the characteristic patterns of amino acid change observed in molecular evolution. These maximum entropy distributions of mutational effects not only provide a tuneable model for molecular evolution, but also have implications for mutational effect prediction and protein engineering.
Keywords: amino acids; deep mutational scanning; epistasis; molecular evolution; protein design.
Conflict of interest statement
Author Declaration The authors declare that they have no conflicts of interest.
Figures
References
-
- Zuckerkandl E, Pauling L, Evolutionary Divergence and Convergence in Proteins, eds. V Bryson H Vogel. (Academic Press, New York: ), (1965).
-
- Eck R, Dayhoff M, Atlas of Protein Sequence and Structure. (National Biomedical Research Foundation, Silver Spring, MD: ), (1966).
-
- Sneath P, Relations between chemical structures and biological activity in peptides. J. Theor. Biol. 12, 157 (1966). - PubMed
-
- Epstein CJ, Non-randomness of amino-acid changes in the evolution of homologous proteins. Nature 215, 355–9 (1967). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
