Identification of a potential sialylation-related pattern for the prediction of prognosis and immunotherapy response in small cell lung cancer
- PMID: 39465788
- PMCID: PMC11479454
- DOI: 10.1097/MD.0000000000040060
Identification of a potential sialylation-related pattern for the prediction of prognosis and immunotherapy response in small cell lung cancer
Abstract
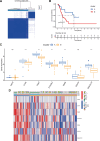
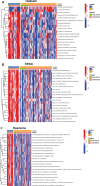
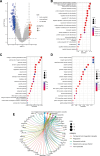
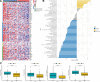
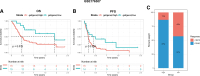
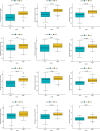
Our study aimed to establish a novel system for quantifying sialylation patterns and comprehensively analyze their relationship with immune cell infiltration (ICI) characterization, prognosis, and therapeutic sensitivity in small cell lung cancer (SCLC). We conducted a thorough assessment of the sialylation patterns in 100 patients diagnosed with SCLC. Our primary focus was on analyzing the expression levels of 7 prognostic sialylation-related genes. To evaluate and quantify these sialylation patterns, we devised a sialylation score (SS) using principal component analysis algorithms. Prognostic value and therapeutic sensitivities were then evaluated using multiple methods. The GSE176307 was used to verify the predictive ability of SS for immunotherapy. Our study identified 2 distinct clusters based on sialylation patterns. Sialylation cluster B exhibited a lower level of induced ICI therapy and immune-related signaling enrichment, which was associated with a poorer prognosis. Furthermore, there were significant differences in prognosis, response to targeted inhibitors, and immunotherapy between the high and low SS groups. Patients with high SS were characterized by decreased immune cell infiltration, chemokine and immune checkpoint expression, and poorer response to immunotherapy, while the low SS group was more likely to benefit from immunotherapy. This work showed that the evaluation of sialylation subtypes will help to gain insight into the heterogeneity of SCLC. The quantification of sialylation patterns played a non-negligible role in the prediction of ICI characterization, prognosis and individualized therapy strategies.
Copyright © 2024 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no funding and conflicts of interest to disclose.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Wang WZ, Shulman A, Amann JM, Carbone DP, Tsichlis PN. Small cell lung cancer: subtypes and therapeutic implications. Semin Cancer Biol. 2022;86(Pt 2):543–54. - PubMed
-
- Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379:2220–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

